How to use
BacARscan scans protein sequences as well as gene sequences provided in fasta format. User(s) have option to cut and paste sequences in the text box or to directly upload the sequence file. Users need to select from the drop box (protein/gene) sequence accordingly. User have an option to select ARG profile HMMs (protein ARG HMM i.e. pARGhmm or Nucleotide ARG HMM i.e. nARGhmm). If user submits gene sequence and select pARGhmm then the query will be first translated to protein sequences and then scanning will be proceeded. Users have an option to scan directly gene query without translating into protein using nARGhmm. For protein sequences users has to select directly protein and pARGhmm from drop box (protein) & (pARGhmm). BacARscan can process upto 10 sequences at a time. If user submits more than 10 sequences, BacARscan will predict only first 10 sequences. Following Figure 1 is an example showing how to submit a query in BacARscan.
Example Sequence(s)
Protein sequence:
>ZP_02959935 hypothetical protein PROSTU_01837 [Providencia stuartii ATCC 25827]
MGIEYRSLHTSQLTLSEKEALYDLLIEGFEGDEAMVVEQSYRRQG
IIQRHMALDNTPISVGYVEAMVVEQSYRRQGIEAMVVEQSYRRQG
KLYHSVGWQIWKGKLFELKQGSYIRSIEEEGGVEAMVVEQSYRRQG
Gene sequence:
>NC_020086.1:c60521-59865 Escherichia coli plasmid pE66An
ATGGCACGGCTGAACAGAGAATCGGTTATTGATGCGGC
GGCTGACGACCCGCAAGCTGGCGCAGAAGCTGGGAATA
TAAACGGGCGTTACTGGATGCGCTGGCGGTGGAGATCT
GCGGGGGAATCCTGGCAGTCATTTCTGCGCAATAATGC
Scan Result
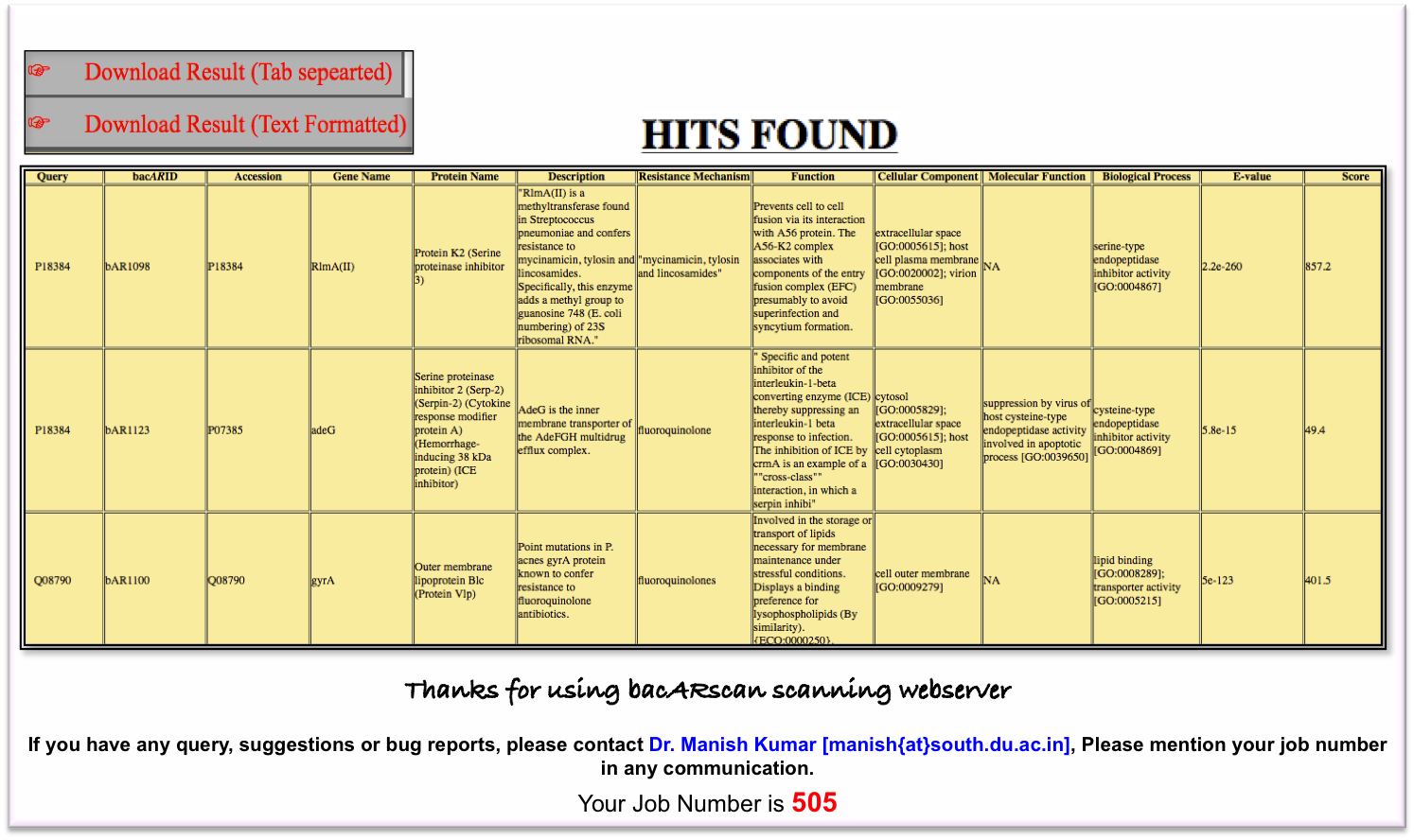
In response to the submitted protein or gene sequence(s), BacARscan generates the scanning result in tabular form. The displayed result contains similarity score and E-value of query against antibiotics resistance genes and also provides detailed annotation of query (Protein accession number, Gene Name, Description, EC number, Organism names, Interactions, 3-D structure information, Fuction, Gene Ontologies, Resistance Mechanism etc). The result can be downloaded in text format as well as in tab separated format. Here is a sample of scanning done in response to three query proteins (Figure 2):