Databases Developed by Group Members
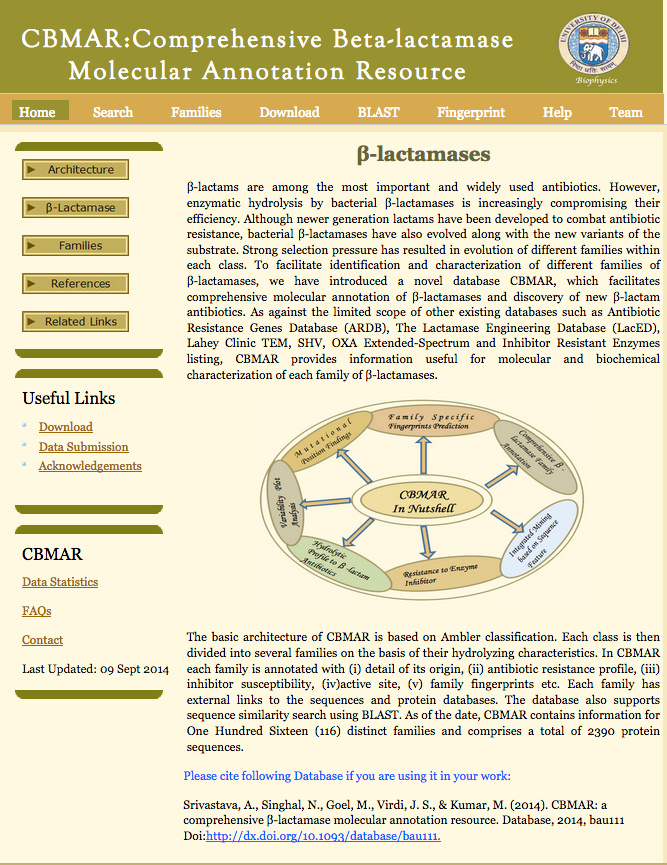
CBMAR To facilitate identification and characterization of different families of β-lactamases, we have introduced a novel database CBMAR, which facilitates comprehensive molecular annotation of β-lactamases and discovery of new β-lactam antibiotics. (Srivastava A, Singhal N, Goel M, Virdi JS and Kumar M. CBMAR: A Comprehensive Beta-Lactamase Molecular Annotation Resource. Database. 2014 Dec 3;2014:bau111. To Access Click Here.
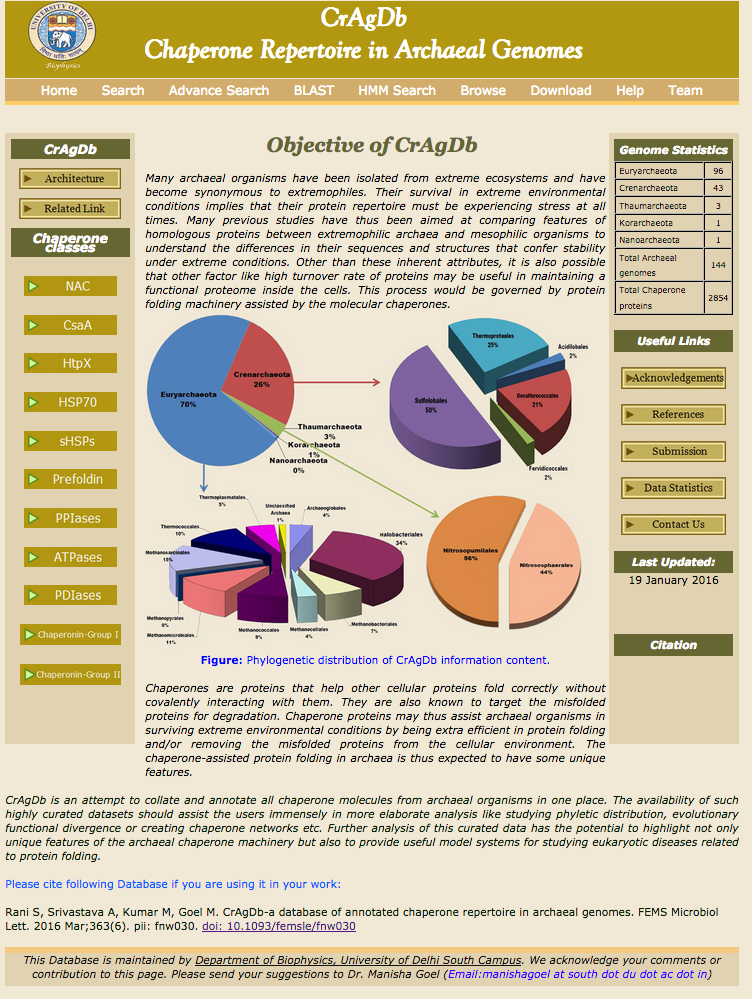
CrAgDb CrAgDb is an attempt to collate and annotate all chaperone molecules from archaeal organisms in one place. The availability of such highly curated datasets should assist the users immensely in more elaborate analysis like studying phyletic distribution, evolutionary functional divergence or creating chaperone networks etc. (Rani S, Srivastava A, Kumar M, Goel M. CrAgDb-a database of annotated chaperone repertoire in archaeal genomes. FEMS Microbiol Lett. 2016 Mar;363(6). pii: fnw030). To Access Click Here.
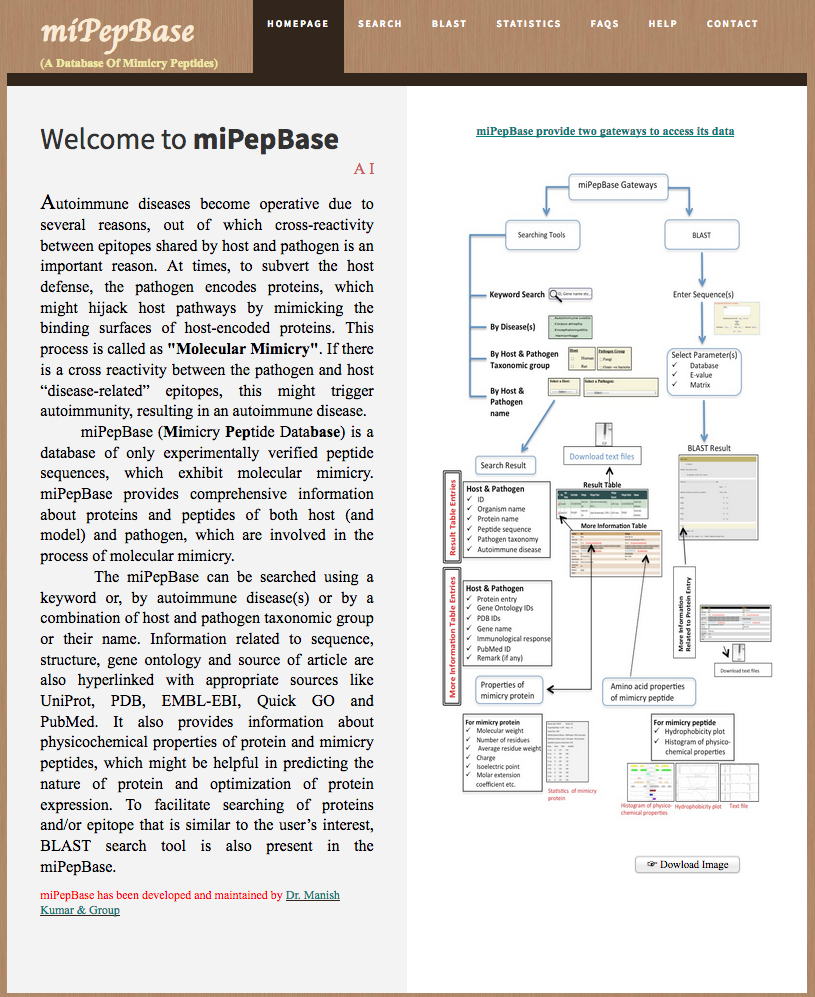
miPepBase miPepBase (Mimicry Peptide Database) is a database of only experimentally verified peptide sequences, which exhibit molecular mimicry. miPepBase provides comprehensive information about proteins and peptides of both host (and model) and pathogen, which are involved in the process of molecular mimicry. (Garg A, Kumari B, Kumar R and Kumar M. miPepBase: A database of experimentally verified peptides involved in molecular mimicry. Frontiers in Microbiology, section Microbial Immunology 8:2053). To Access Click Here.