Help
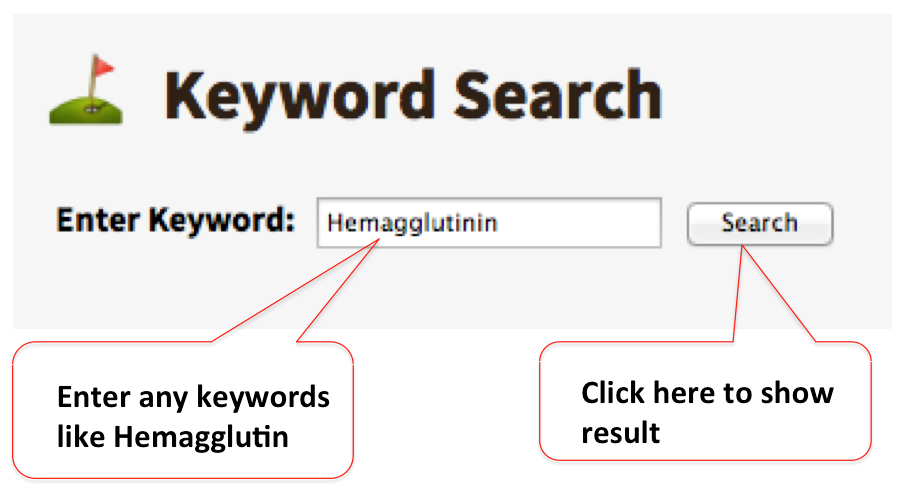
Keyword Search
Advanced Search
Search Result
BLAST
FAQs
A keyword search feature allows users to search the database with keywords such as host name, pathogen name, disease, and ID, etc.
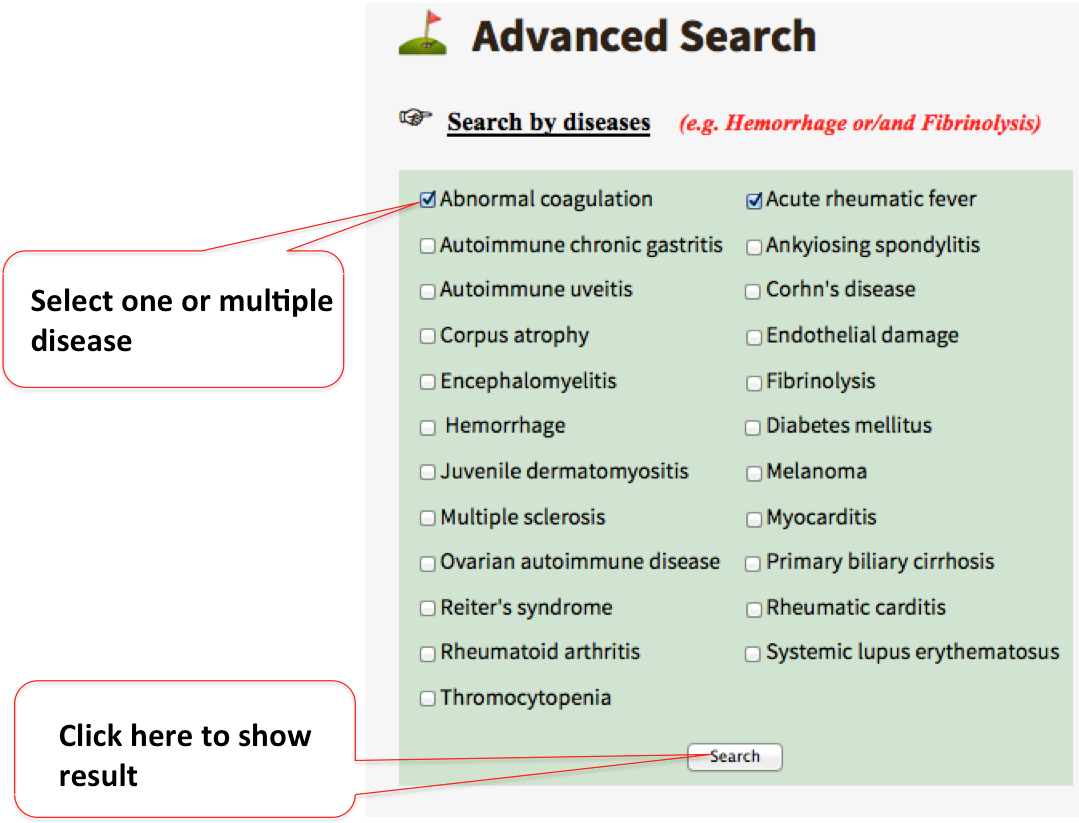
Advanced search option allow users to specify a range within which search will be restricted. It is further divided into three sections:
1.
By diseaseThis allow users to restrict the search by one or multiple disease(s) caused due to molecular mimicry.
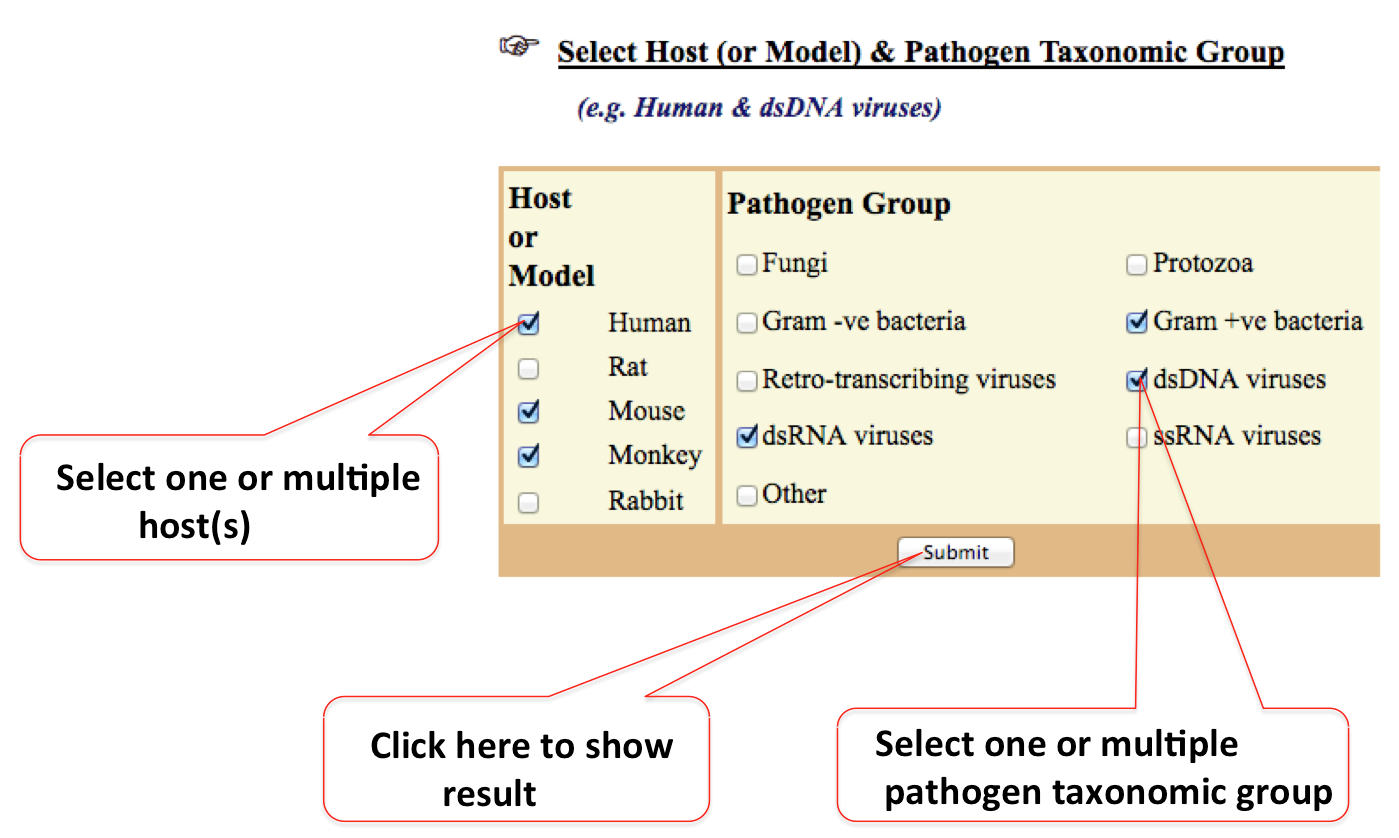
2. By Host & Pathogen Group
In this type of search users can restrict the search to one or multiple host(s) and pathgen taxonomy involved in molecular mimicry. Pathogen are categorized according to their taxonomy like dsDNA, dsRNA, dsRNA and Fungi etc.
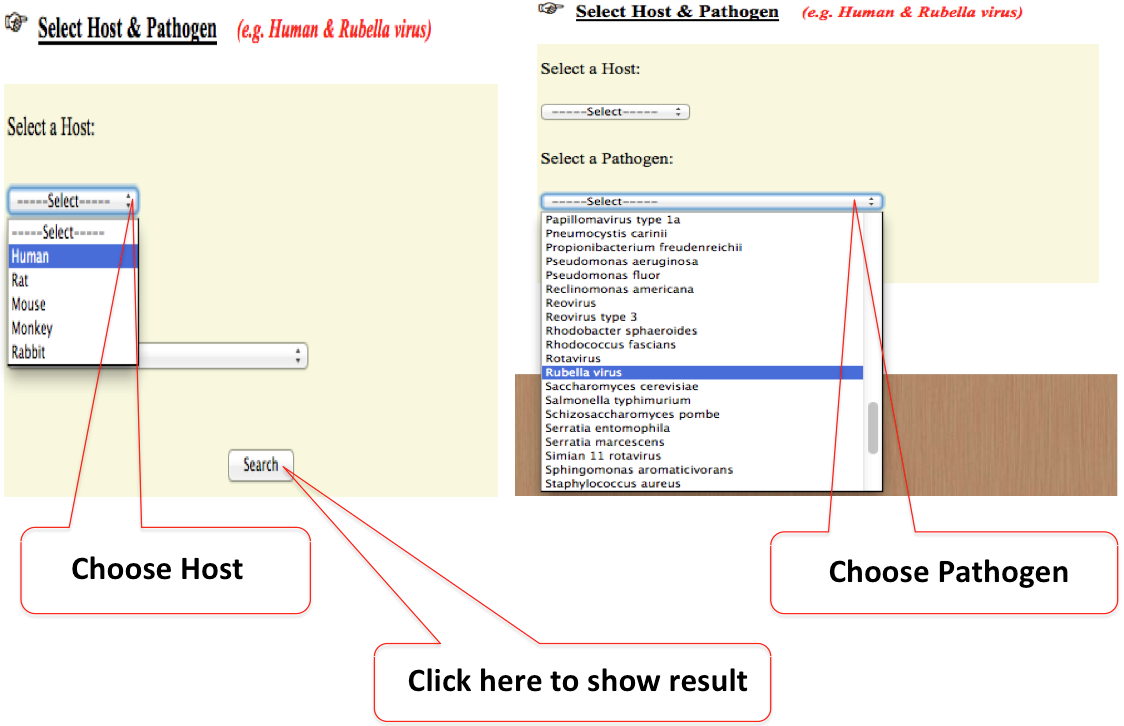
3. Host & Pathogen
Drop down menu of host & pathogens allows users to specify exact pathogenic organism. The following example shows how to restrict the search with host as human & pathogen as Rubella virus.
Search Result
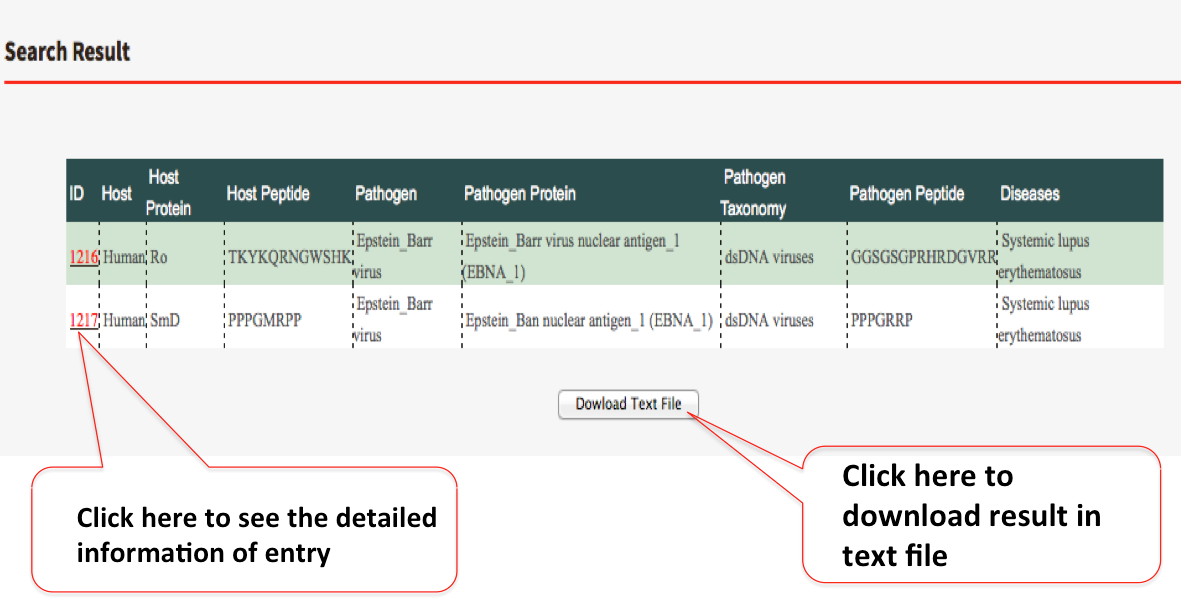
Search by any of the above descibed methods will give result in tablular form which will include ID, host name, host protein name, host peptide sequence, pathogen name, pathogen protein name, pathogen peptide sequence, pathogen taxonomy and disease. The user can also download the whole information in text form. The full detail of each each row can be displayed by the ID (red colored).
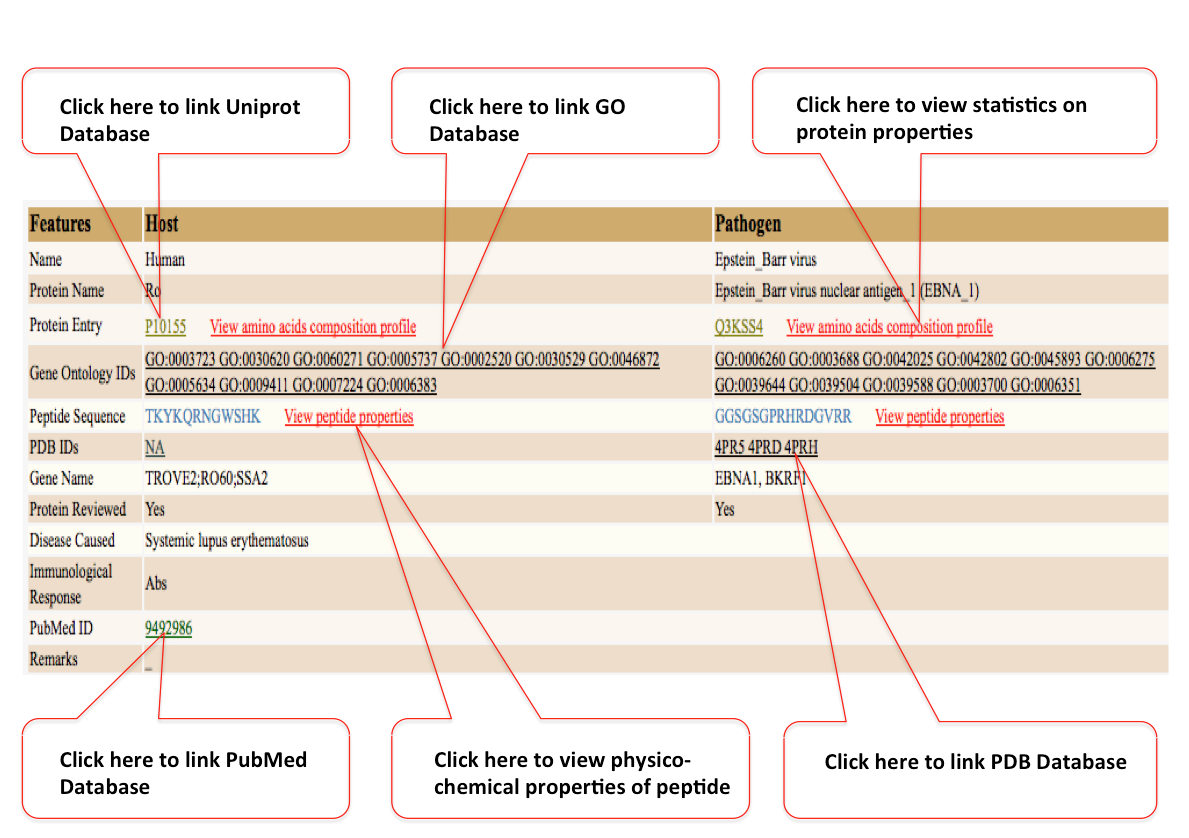
More detailed information page include Go Ids, PDB IDs, Protein review status information, Immunological response (T-cell/Abs) and PubMed ID.
A. View Physico-chemical Properties of Proteins
Users can also get the information of different physico-chemical proterties of Host and Pathogen proteins by clicking
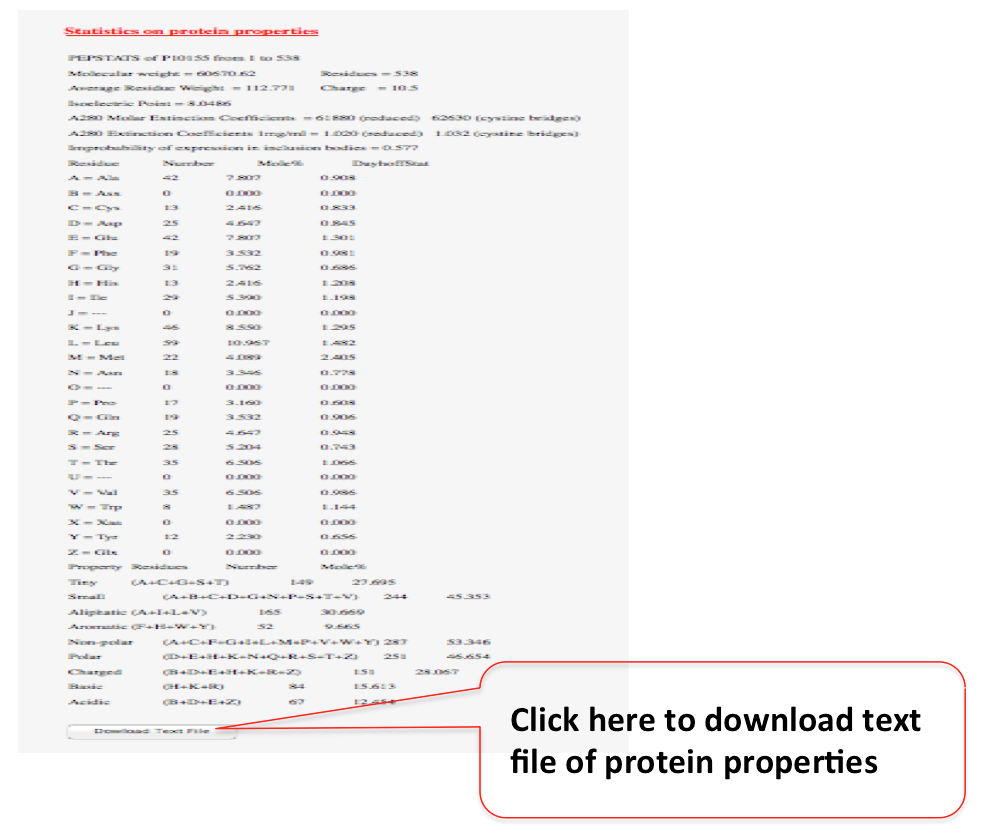
“view amino acid composition profile” (red colored). The properties includes: Molecular weight, Number of residues, Average residue weight, Charge, Isoelectric point, For each type of amino acid: number, molar percent, DayhoffStat, For each physico-chemical class of amino acid: number, Probability of protein expression in E. coli inclusion bodies, Molar extinction coefficient (A280), and Extinction coefficient at 1 mg/ml (A280).This information file can also be downloaded in text format.
B. View physico-Chemical properties of peptides
The physico-chemical properties of peptide sequences of both host and pathogen is also available to the users. To retrive this information click the
“view peptide properties” (red colored) . This also include plots of various amino acid properties namely (i) Hydrophobicity plots, (ii) Histogram of the presence of residues with the physico-chemical properties: Tiny, Small, Aliphatic, Aromatic, Non-polar, Polar, Charged, Positive, and Negative. This whole data and the plot can also be downloaded.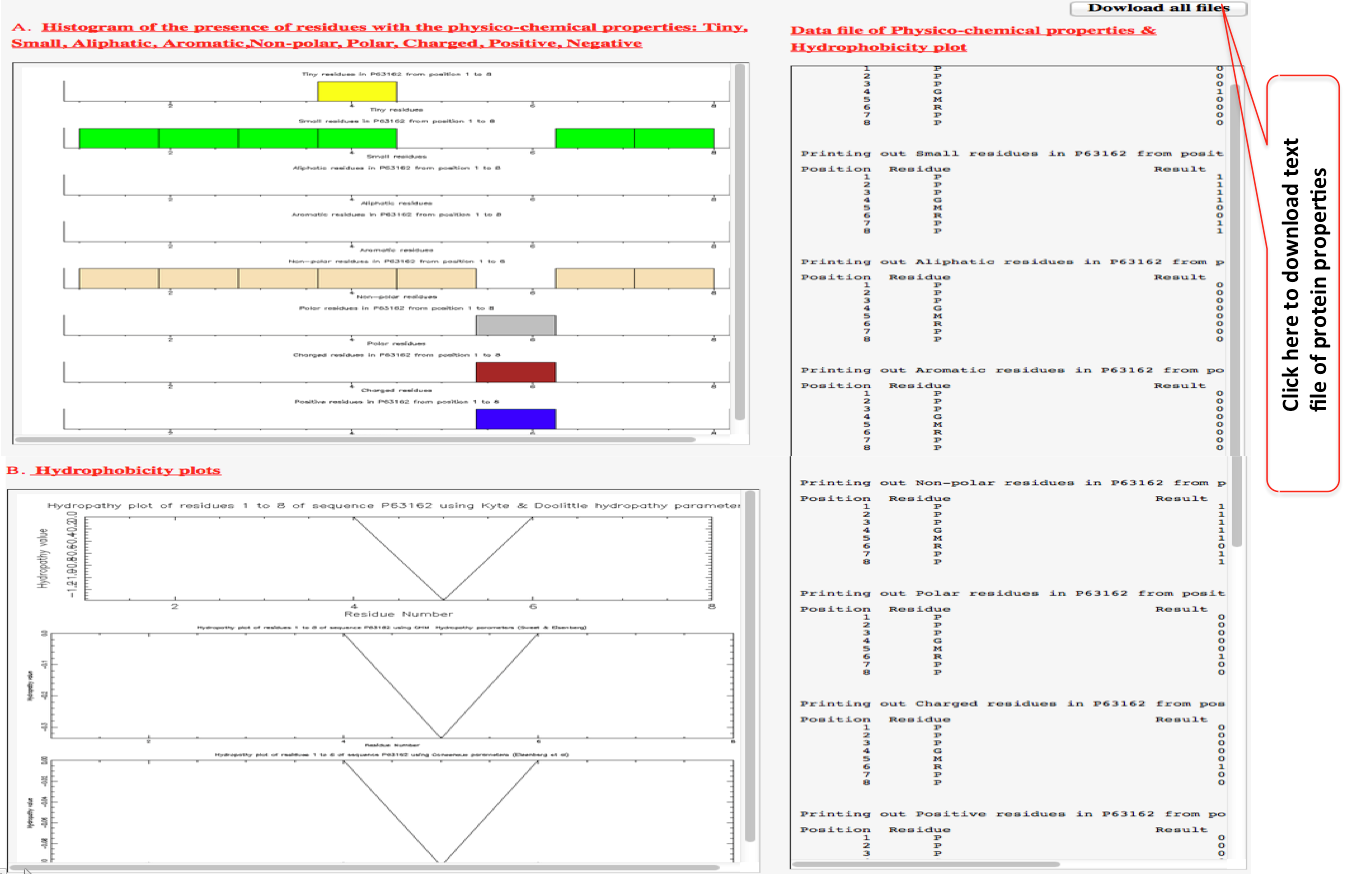
BLAST
(Search for similar sequences)
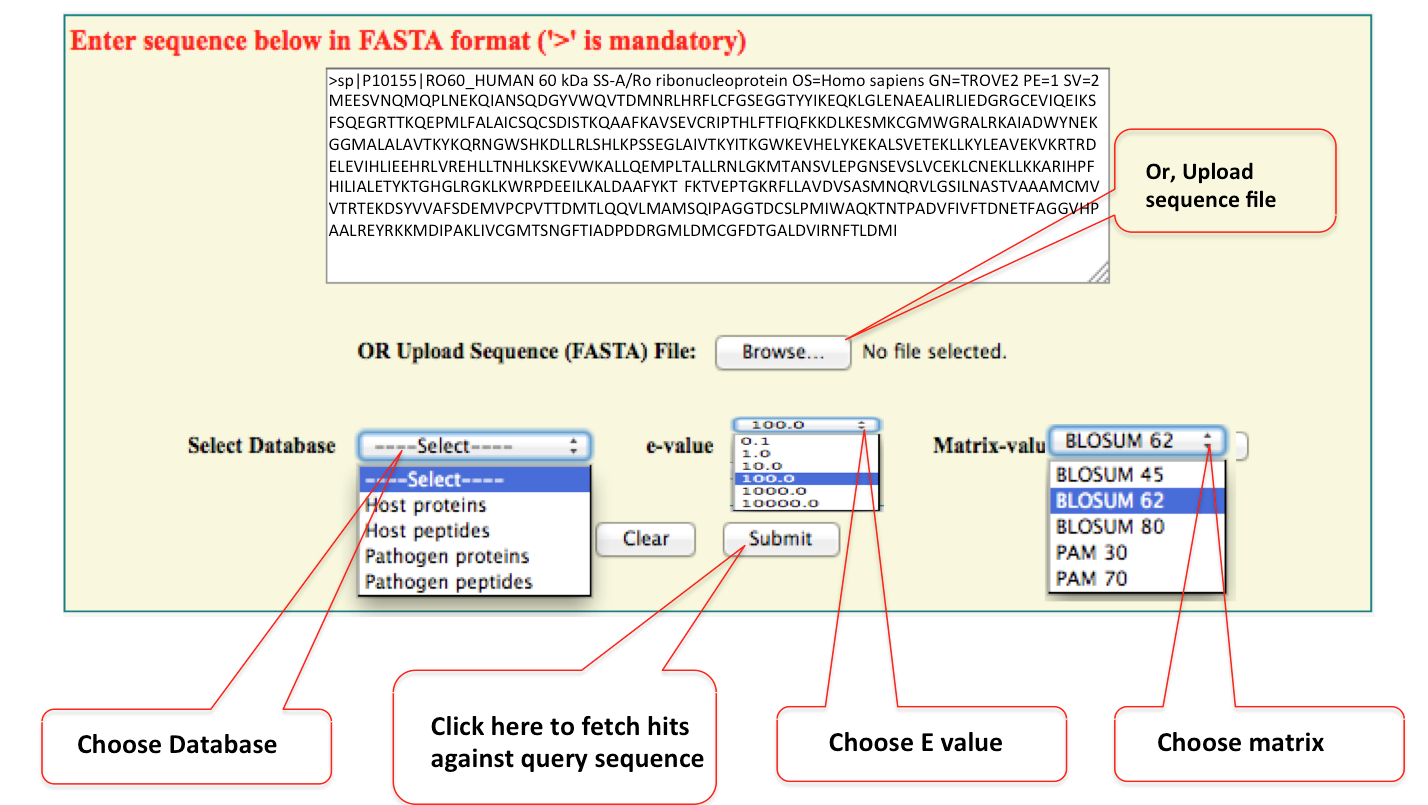
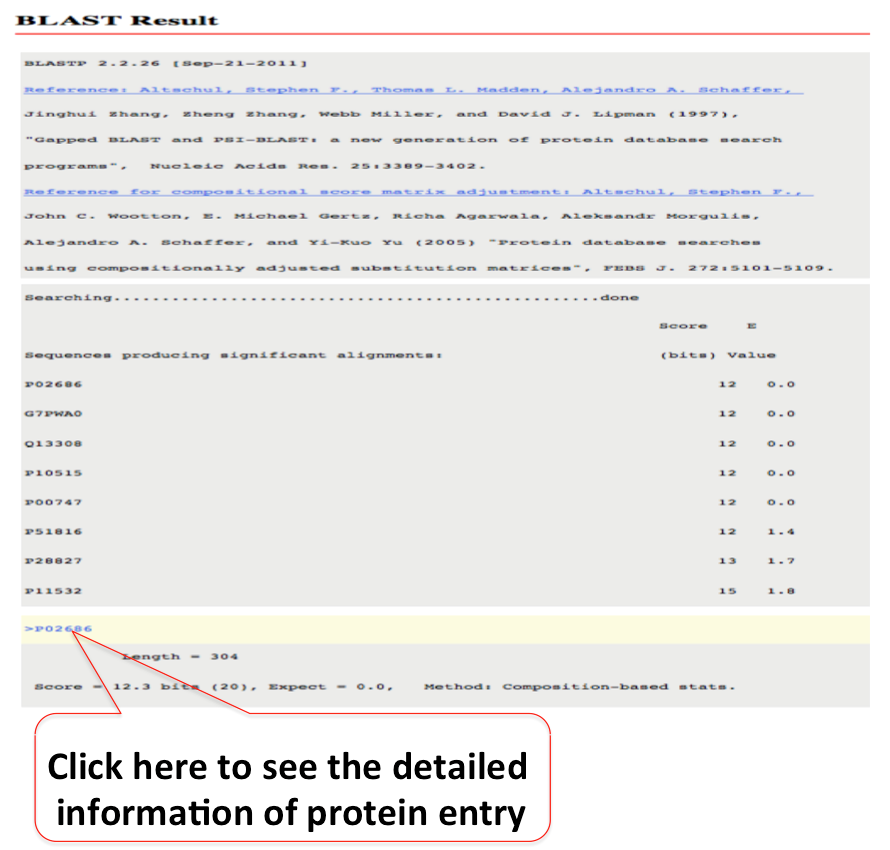
Blast can be used to find similar sequences against of your interest, if it is present in miPepBase. There is option to select different type of databases. The default expectation value (E-value) is 100 and alignment scoring matrix is BLOUSM62. They can be changed using 'drop down options' of E-value and matrix value.
Point to be remember during search of sequenceFor all types of sequence searches:
•Do not enter spaces or dashes. (Example: do not use: M L P K L, M-L-P-K-L)
•Do not enter amino acid modifications, phosphorylation, terminal and other marks. (Example: do not use: Myr-GE(pT)RAPL-NH2)
•Remove the D prefix from D-amino acids. (Example: do not use: Gly-Glu-Thr-Arg-D-Ala-Pro-Leu)
•Do not use three letter amino acid code, or full names. (Example: do not use: Gly-Glu-Thr-Arg-Ala-Pro-Leu)
•Use only single letter amino acid code, uppercase or lowercase. (Example: for all of the above, use: MLPKL, mlpkl)
FAQs
Frequently asked questions (FAQ) page provide list of questions and answers, that supposed to be commonly strike in users mind during using it.
