Welcome to mRNALoc
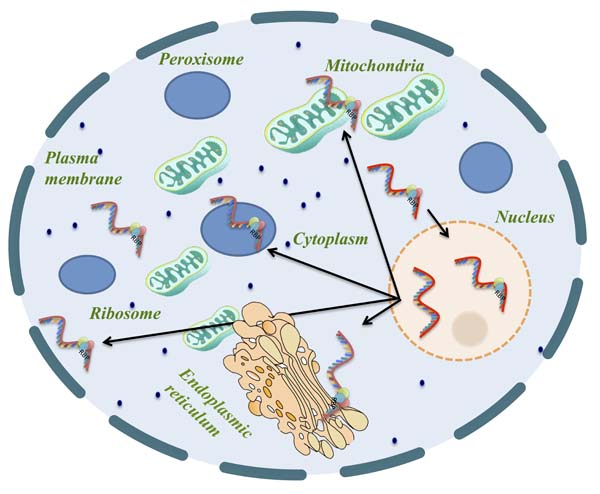
Recent evidence suggests that the localization of mRNAs near the subcellular compartment of the translated proteins is a more robust cellular tool, which optimizes protein expression, post-transcriptionally. Retention of mRNA in the nucleus can regulate the amount of protein translated from each mRNA, thus allowing a tight temporal regulation of translation or buffering of protein levels from bursty transcription. Besides, mRNA localization performs a variety of additional roles like long-distance signaling, facilitating assembly of protein complexes and coordination of developmental processes.
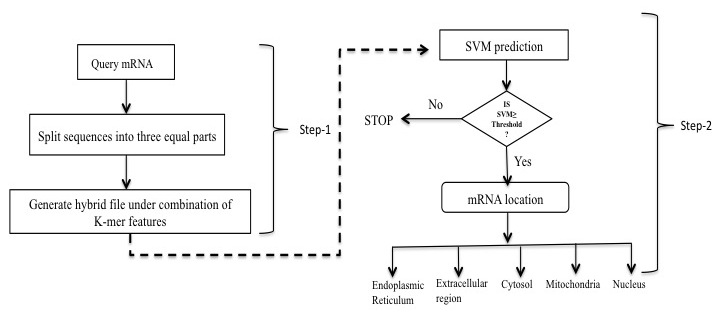
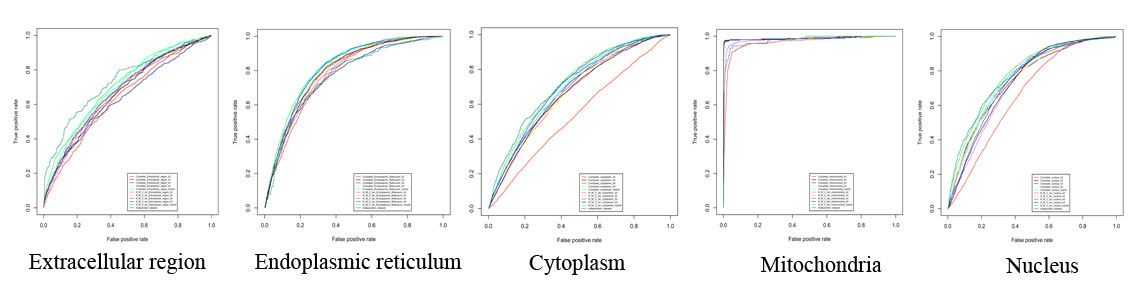
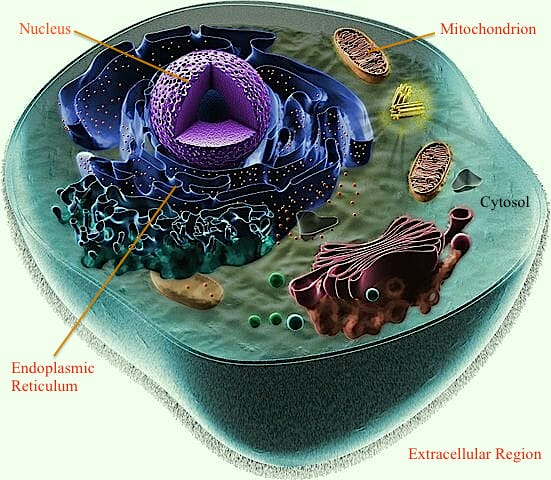
Here we describe a novel machine-learning based tool, mRNALoc, to predict five sub-cellular locations of eukaryotic mRNAs using cDNA/mRNA sequences. During five fold cross-validations, the maximum overall accuracy was 65.19, 75.36, 67.10, 99.70 and 73.59 % for the extracellular region, endoplasmic reticulum, cytoplasm, mitochondria, and nucleus, respectively. Assessment on independent datasets revealed the prediction accuracies of 58.1, 69.23, 64.55, 96.88 and 69.35 % for extracellular region, endoplasmic reticulum, cytoplasm, mitochondria, and nucleus, respectively. The corresponding values of AUC were 0.76, 0.75, 0.70, 0.98 and 0.74 for the extracellular region, endoplasmic reticulum, cytoplasm, mitochondria, and nucleus, respectively. The mRNALoc standalone software and web-server are freely available for academic use under GNU GPL.
mRNALoc predicts
Latest Updates
-
October 19th, 2019
This is our first release of mRNALoc V 1.0
-
Cite US
mRNALoc: a novel machine-learning based in-silico tool to predict mRNA subcellular localization
-
Our Other Predictors
-
Reach US