SubMitoPred
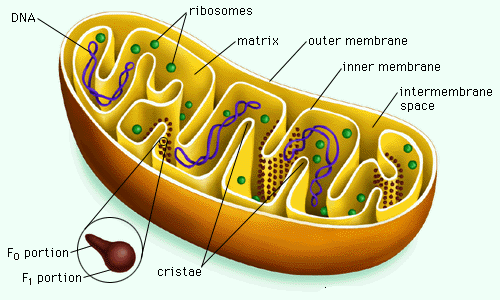
SubMitoPred is support vector machine (SVM) and Pfam based two level predictor which predict proteins localize to mitochondrial as well as submitochondrial location. This predictor use amino acid composition of 25 residues of N-terminal amino acid and the remaining amino acid as SVM input. So here the total length of input vector is 40. At first level it discriminate mitochondrial protein from non-mitochondrial and at second level it predict sub-mitochondrial locations. Here we consider four different submitochondrial location: (i) Inner membrane (ii) Outer membrane (iii) Matrix and (iv)Inter membrane space. Here we found maximum accuracy 94.21% at first level and 76.90% accuracy in inner membrane, 83.80% in outer membrane, 82.98% in inter membrane space and 83.86% accuracy in matrix at second level.