How it Works
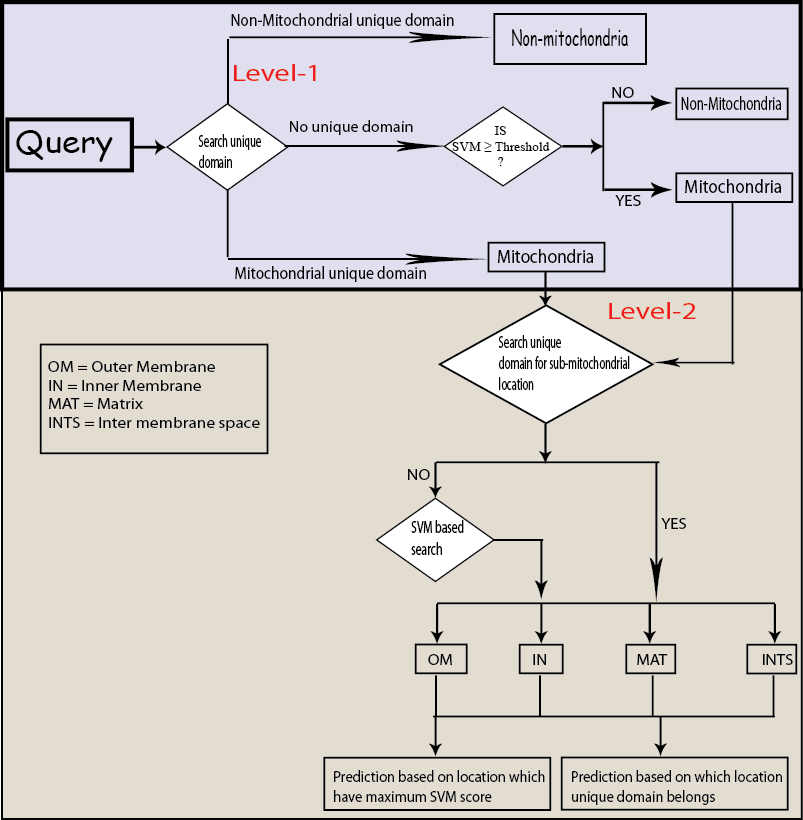
SumMitoPred is a combined method of Pfam domain information and support vector machine (SVM) based predictor. In this method, first our predictor search unique Pfam domain for mitochondrial and non-mitochondrial proteins at first level, if it found non-mitochondrial unique domain, it predict query as non-mitochondria and if it found mitochondrial unique domains, it moves to second level. Here one more condition is that if it does not found any unique domain, it calculate N-terminal and remaining amino acid composition and predict according to SVM score (if SVM score is less than threshold, predict as non-mitochondria if SVM score greater than threshold, predict as mitochondria and move to second level).
At second level, it again search unique domain for sub-mitochondrial location, if present, it predict the same location from where the unique domain belongs and if not, it predict location which have maximum SVM score. Figure 1 shows the overall architecture of SubMitoPred.
At second level, it again search unique domain for sub-mitochondrial location, if present, it predict the same location from where the unique domain belongs and if not, it predict location which have maximum SVM score. Figure 1 shows the overall architecture of SubMitoPred.