CrAgDb
Chaperone Repertoire in Archaeal Genomes

CsaA System
The CsaA protein was first characterized in Bacillus subtilis as a chaperone of 15 kDa, which has an export related activity (Ref 1, Ref 2, Ref 3). CsaA is one of the few chaperone proteins that are shared between bacteria and archaea but are absent in eukaryotes. It has been suggested that the proteins CsaA and SecB have overlapping functions, although these proteins share no sequence identity or structural similarity (Ref 4). CsaA protein from B.subtilis has the ability to bind with SecA subunit of E.coli and a number of preproteins for translocation. During the translocation pathway, CsaA may specifically bind to the long hydrophobic segments of preproteins or may hide the exposed hydrophobic patches of denatured proteins to prevent the aggregation of unfolded cytoplasmic proteins. CsaA appears to have higher affinity for the denatured proteins than the native ones.
The crystal structure of CsaA protein from Thermus thermophilum revealed that it is a homodimer, each monomer consists of a β-barrel like structure which is similar to an oligonucleotide binding fold (OB) and the dimer interface is formed by the short N and C terminal extensions from the β-barrel core region (Ref 5). Several lines of evidence associate CsaA with the C-terminal domain of methionyl-tRNA synthetases and its nearest structural relative Trbp111, and they are considered to be members of the same family (Ref 6; Ref 7). All the three members of the family share significant similarity at the sequence and structural level. All the three proteins were shown to form dimers and the residues responsible for the dimerization are highly conserved (Ref 8 ; Ref 9). Structure based sequence analysis suggest that CsaA may also be a tRNA binding protein, discovered after Trbp111, which may act as a tRNA specific chaperone protein helping to maintain the tRNA native folds. However, the function of CsaA protein is still unclear, as it may have only an export related function with replacement of ancestral tRNA binding activity.
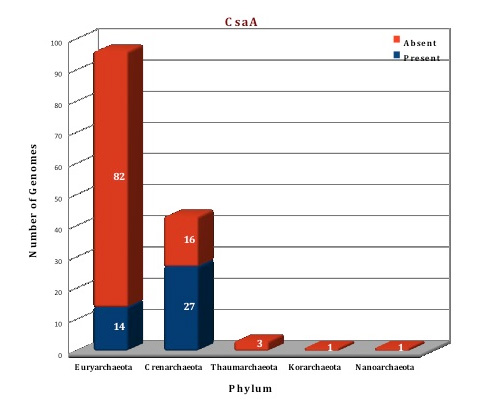
From the analysis of our database, CsaA homologous are found in only 41 archaeal genomes. CsaA protein is totally absent from the phylums Korarchaeota, Nanoarchaeota and Thaumarchaeota. It is also absent from many orders of Euryarchaeota (Archaeoglobales, Methanobacteriales, Methanocellales, Methanosarcinales, Methanomicrobiales, Methanococcales, Methanopyrales, Thermococcales) and from the order Thermoproteales of phylum Crenarchaeota. Four bacterial structures of CsaA protein are available from B.subtilis (2NZH) (Ref 10), T.thermophilus (1GD7), B.anthracis (3G48), and A.tumefaciens (2Q2I). Although, homologs of CsaA are found in some archaea, no structure is available from archaeal organisms. We have modelled the structure of CsaA from Picrophilus torridus by using the template from B.subtilis.
|
|
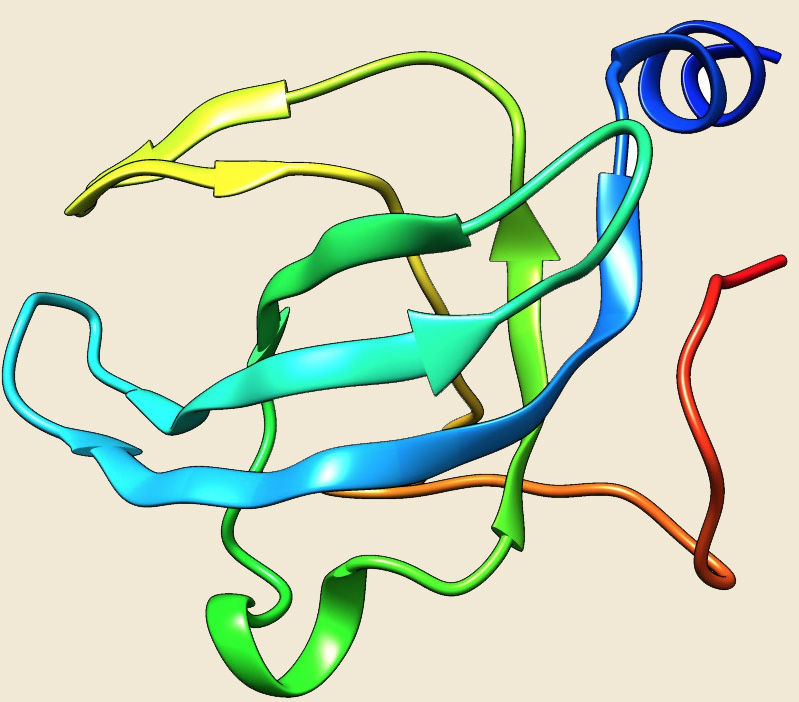
Figure: The modeled structure of CsaA from Picrophilus torridus. |
|
|
|
Figure: Distribution of CsaA chaperone proteins in 144 archaeal genomes. |
 |
References:
1. Suppression of the growth and export defects of an Escherichia coli secA(Ts) mutant by a gene cloned from Bacillus subtilis.![]()
2. Chaperone-like activities of the CsaA protein of Bacillus subtilis.![]()
3. Interaction of Bacillus subtilis CsaA with SecA and precursor proteins.![]()
4. Crystal structure of the bacterial protein export chaperone secB.![]()
5. The crystal structure of the ttCsaA protein: an export-related chaperone from Thermus thermophilus.![]()
6. Crystal structure of trbp111: a structure-specific tRNA-binding protein.![]()
7. A recurrent general RNA binding domain appended to plant methionyl-tRNA synthetase acts as a cis-acting cofactor for aminoacylation.![]()
8. Functions of isolated domains of methionyl-tRNA synthetase from an extreme thermophile, Thermus thermophilus HB8.![]()
9. Structure-specific tRNA-binding protein from the extreme thermophile Aquifex aeolicus.![]()
10. Crystallographic analysis of Bacillus subtilis CsaA.![]()
11. Phage display and crystallographic analysis reveals potential substrate/binding site interactions in the protein secretion chaperone CsaA from Agrobacterium tumefaciens.![]()
