CrAgDb
Chaperone Repertoire in Archaeal Genomes

Many archaeal organisms have been isolated from extreme ecosystems and have become synonymous to extremophiles. Their survival in extreme environmental conditions implies that their protein repertoire must be experiencing stress at all times. Many previous studies have thus been aimed at comparing features of homologous proteins between extremophilic archaea and mesophilic organisms to understand the differences in their sequences and structures that confer stability under extreme conditions. Other than these inherent attributes, it is also possible that other factor like high turnover rate of proteins may be useful in maintaining a functional proteome inside the cells. This process would be governed by protein folding machinery assisted by the molecular chaperones.
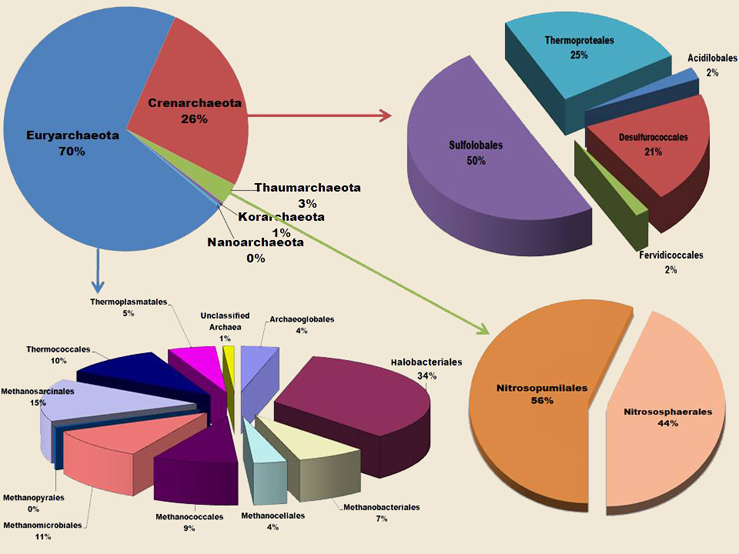
Figure: Phylogenetic distribution of CrAgDb information content.
Chaperones are proteins that help other cellular proteins fold correctly without covalently interacting with them. They are also known to target the misfolded proteins for degradation. Chaperone proteins may thus assist archaeal organisms in surviving extreme environmental conditions by being extra efficient in protein folding and/or removing the misfolded proteins from the cellular environment. The chaperone-assisted protein folding in archaea is thus expected to have some unique features.
CrAgDb is an attempt to collate and annotate all chaperone molecules from archaeal organisms in one place. The availability of such highly curated datasets should assist the users immensely in more elaborate analysis like studying phyletic distribution, evolutionary functional divergence or creating chaperone networks etc. Further analysis of this curated data has the potential to highlight not only unique features of the archaeal chaperone machinery but also to provide useful model systems for studying eukaryotic diseases related to protein folding.
Please cite following Database if you are using it in your work:
Rani S, Srivastava A, Kumar M, Goel M. CrAgDb-a database of annotated chaperone repertoire in archaeal genomes. FEMS Microbiol Lett. 2016 Mar;363(6). pii: fnw030. doi: 10.1093/femsle/fnw030