CrAgDb
Chaperone Repertoire in Archaeal Genomes

Information & Help
CrAgDb is a collection of manually curated Chaperone classes and their Families. All data in this database is collected after extensive manual literature search and review of other publicly available general databases like NCBI, UniProt. Currently, the database incorporates nearly 2843 entries for 10 different classes and their families. It is expected that the number of entries would increase once the database becomes available to world-wide use.
Table of Contents:
| Architecture | Related Fields | Keyword Search | Advance Search |
| BLAST Search | HMM Search | Browse | Submission and Upgradation of CrAgDb |
| Download Information |
Architecture |
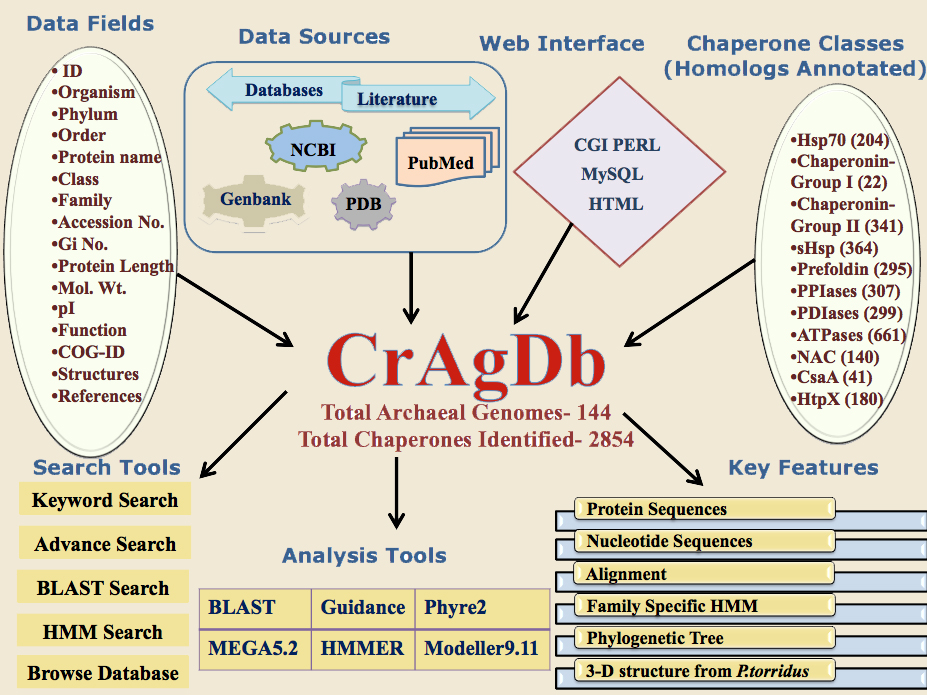 |
Related Fields |
Data Management at CragDb:
The basic structure of a data unit is given below:
| Field Name | Description of Field |
| ID: | CrAgDb id is designed as a five digit unique identifier assigned to each entry of the database. By using these unique identifiers, the database entries can be searched. |
| Organism: | This field have information about 144 sequenced archaeal genomes from which the chaperone data is analyzed. The field contains the scientific names for archaeal organisms with their strains. |
| Protein Name: | This name is representing the standard name of the chaperone protein found in database record. |
| Accession No. : | The entry contains the unique accession number of NCBI for each sequence |
| GI: | Gene identifier is the unique number given to each protein sequence by GenBank. |
| Length: | Contains the amino acid length of chaperone protein. |
| Molecular Weight: | Molecular weight of the chaperone proteins in Daltons (Da). |
| pI: | Isoelectic point of the each and every chaperone protein. |
| Function: | A short description of the function of that protein in the cell. |
| COG/arCOG ID: | Cluster of Orthologous groups represent the ID provided by COG database to each class of the Chaperone. arCOGID is specifically related to Archaeal organisms only. |
| Related Structures: | This field have the information about the available structures for each class and family from archaea, bacteria and eukaryotes. The entry is linked with PDB database via PDBID and its related literature is linked with PubMed through PubMedID. |
| References: | The published references of archaeal genomes and references for each class are specified under the reference section and at the end of the page. |
| Keyword Search: | Keyword Search is the easiest way to search for a particular chaperone of interest. In the search field there are two options, containing search which will search anyword in the phrase and the exact search will search for exact word match. |
| CrAgDb-BLAST: | Alignment tool to search for homologous chaperone proteins present in CrAgDb. The users can either paste the sequence in fasta format through the input window provided or browse the sequence from the disk. |
This is a very simple and easy search option. User can search any query in any field such as class (e.g. PPIases), family (e.g. Cyclophilin), Order (e.g. Desulfurococcales) , Phylum (e.g.Crenarchaeota), Function (e.g. Accelerates folding of proteins). Results will be displayed according to the selected fields to be displayed. e.g. class, COGID etc.
How to search the database?
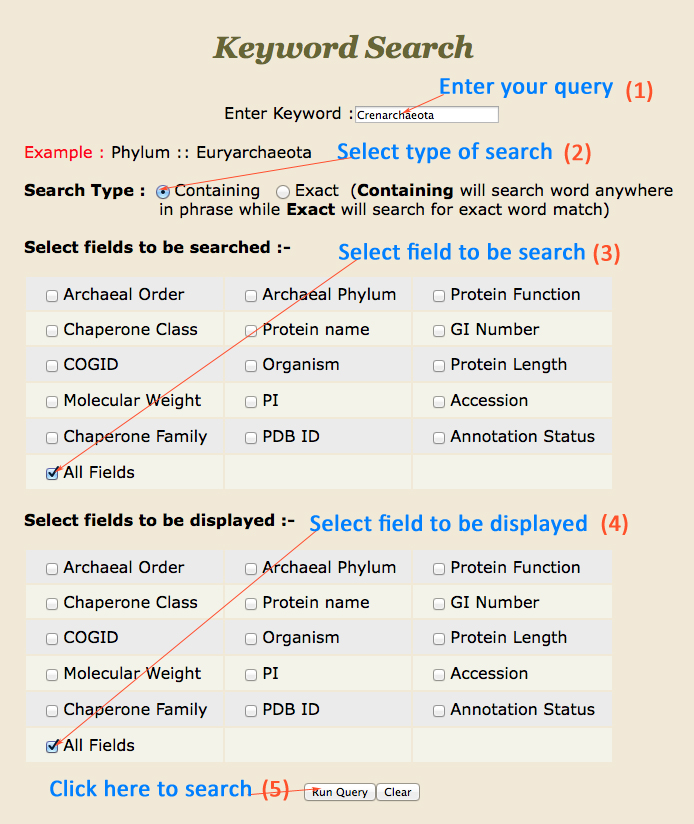 |
Search Result :
User can restrict fields to be displayed in search result. All checked fields are displayed in result. If user did not select any checkbox then default fields will be displayed. The result shows no record when relevant data is not found in database.
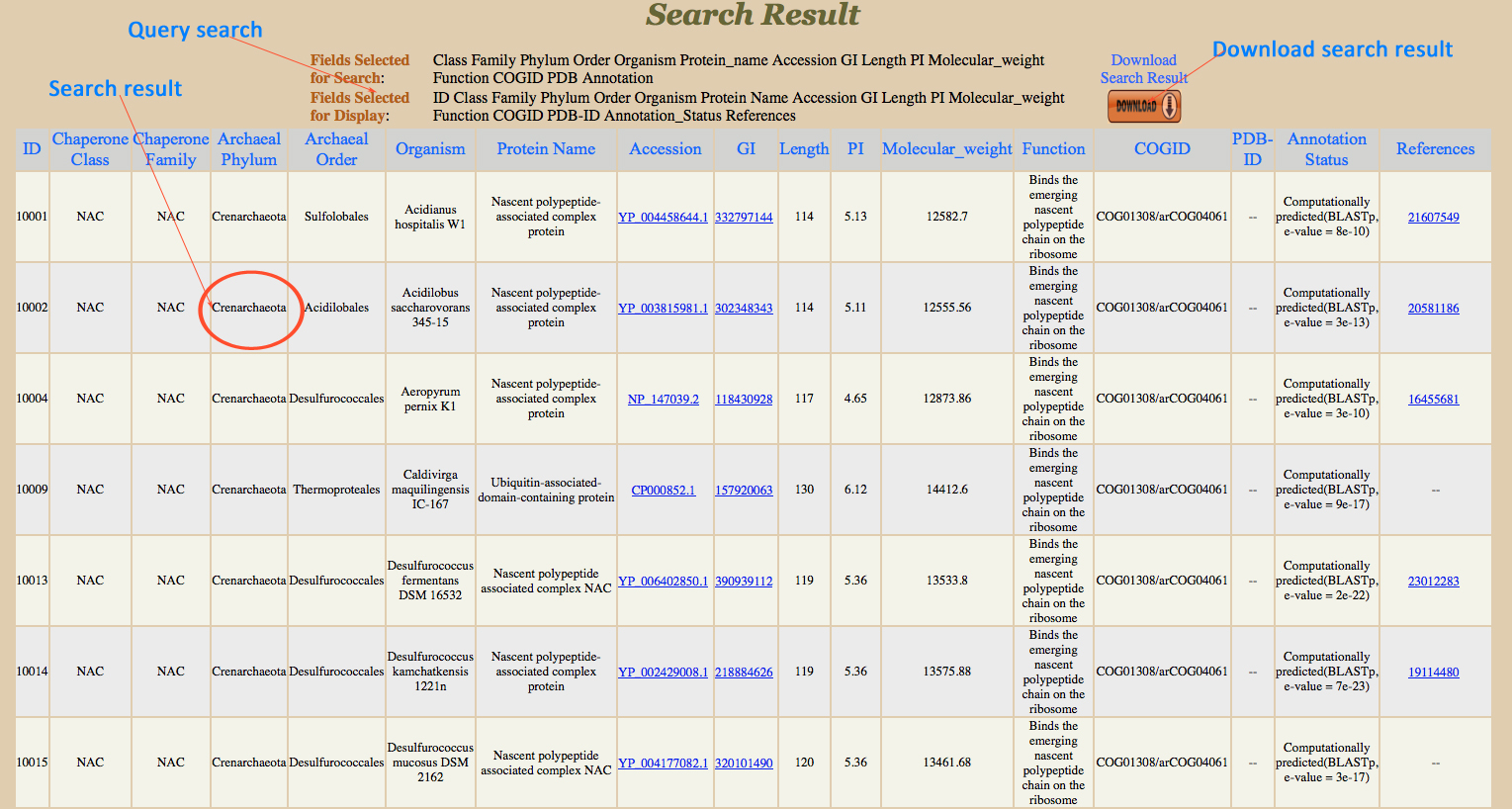 |
Advance Search |
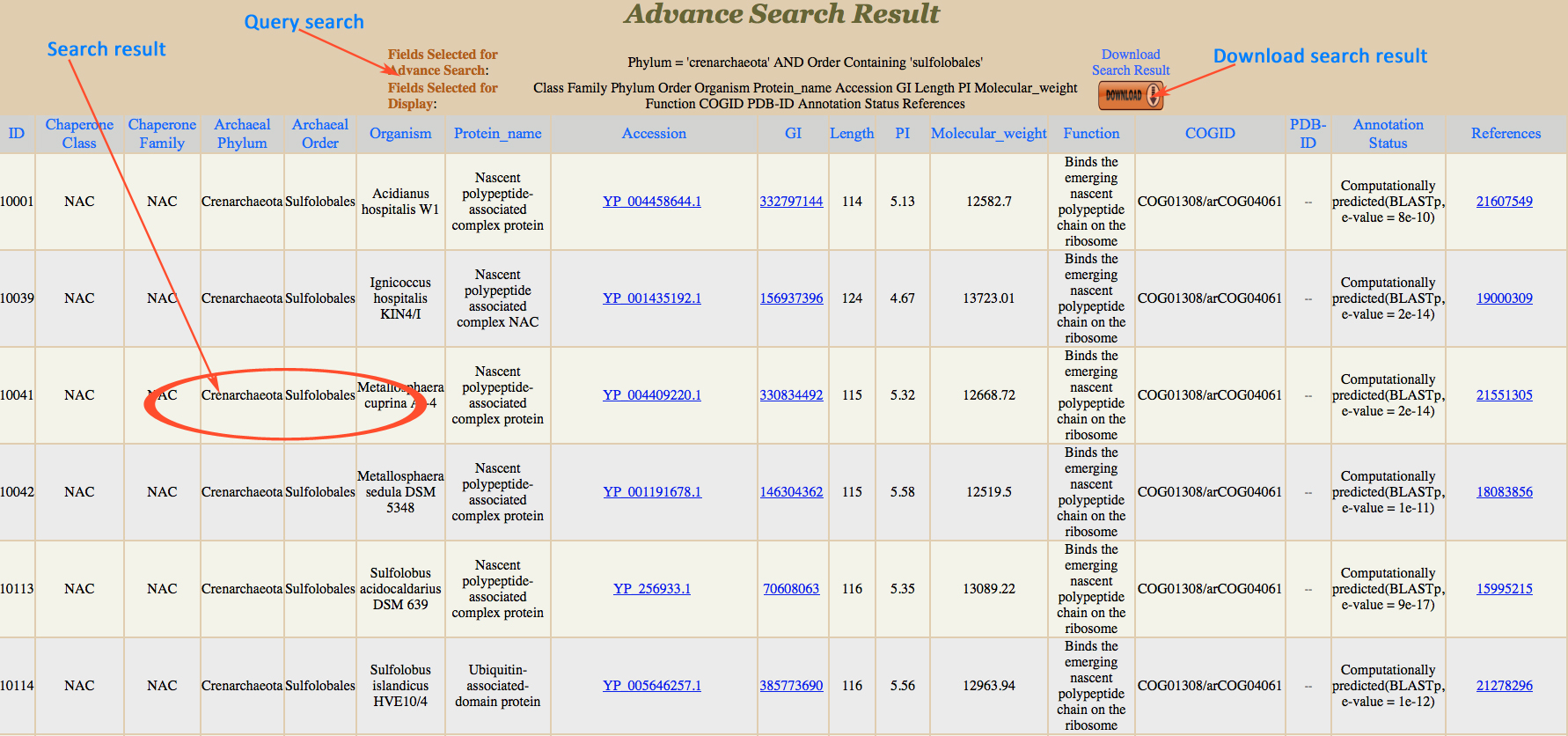
This search has multiple options which give very refined result for the user. Here user has been provided with different search options Field name, Match option (exact, containing, not equal to). Keyword column is provided to add input from user e.g. Crenarchaeota, Sulfolobales, etc. and then user can apply conditions (AND & OR). For example:-
1. If user is giving Archeal Phylum in the field name and put value like crenarchaeota,the user will opt for (exact) match, result page will display all the fields belongs to crenarchaeota class.
2. If condition AND is applied then another search field (e.g. Archeal Order) will be opened, user can put value like Sulfolobales, then results with crenarchaeota containing sulfolobales in the field selected will be displayed.
3. If condition OR with the above input, the user will get all fields containing crenarchaeota and all of them will have an order field sulfolobales.
*Note: While submitting, the advanced search for various conditons, the last search box in the conditions must be blank. By pressing search, desired result will be displayed.
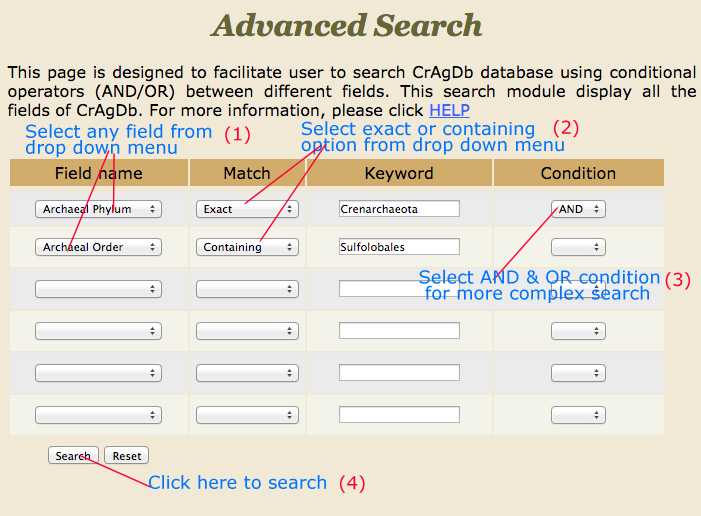 |
 |
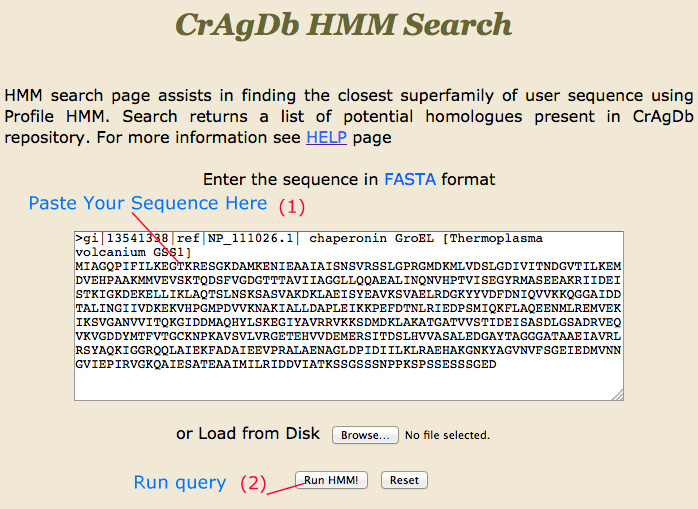
HMM Search |
HMM-Search page assists the user to search against the CrAgDb database with a profile HMM. Output ranked lists of the sequences with the most significant matches to the profile.
 |
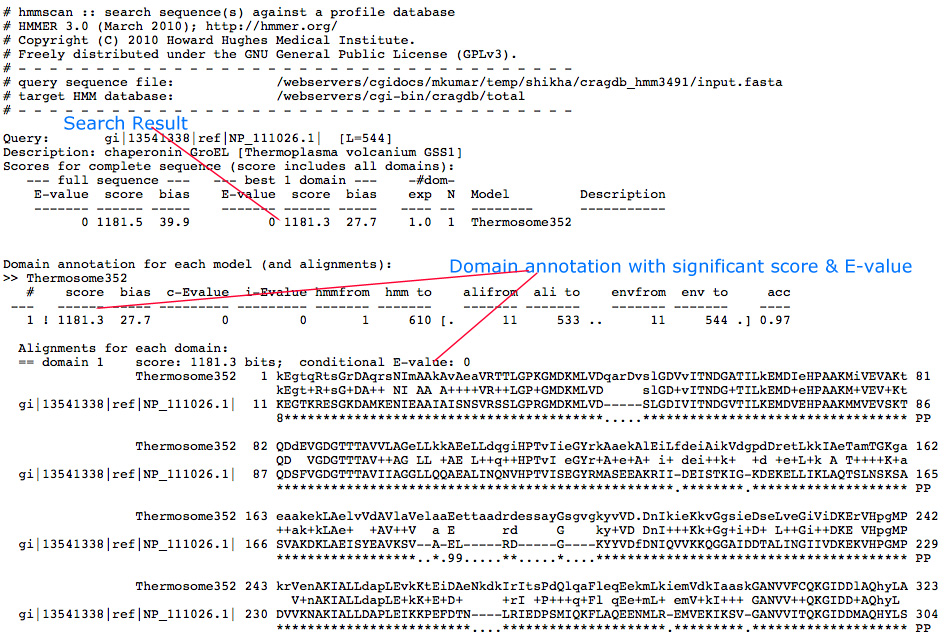
HMM Search Result
 |
How to browse the database ?
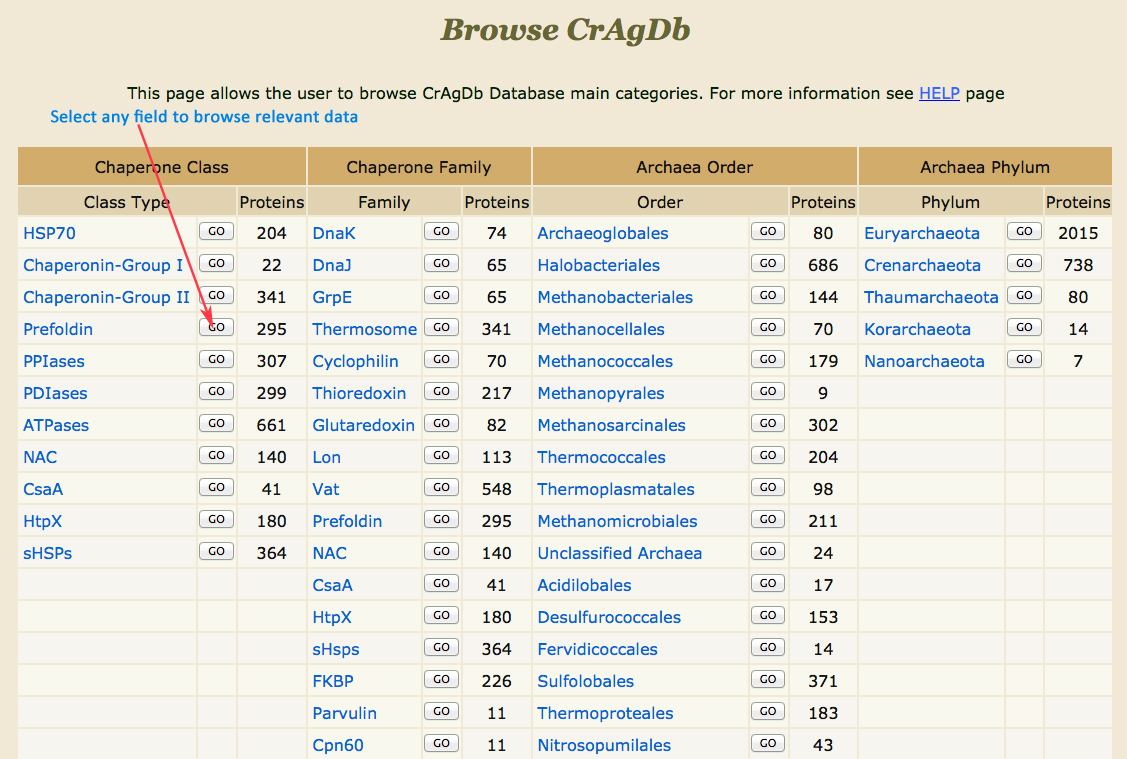 |
Browse Result
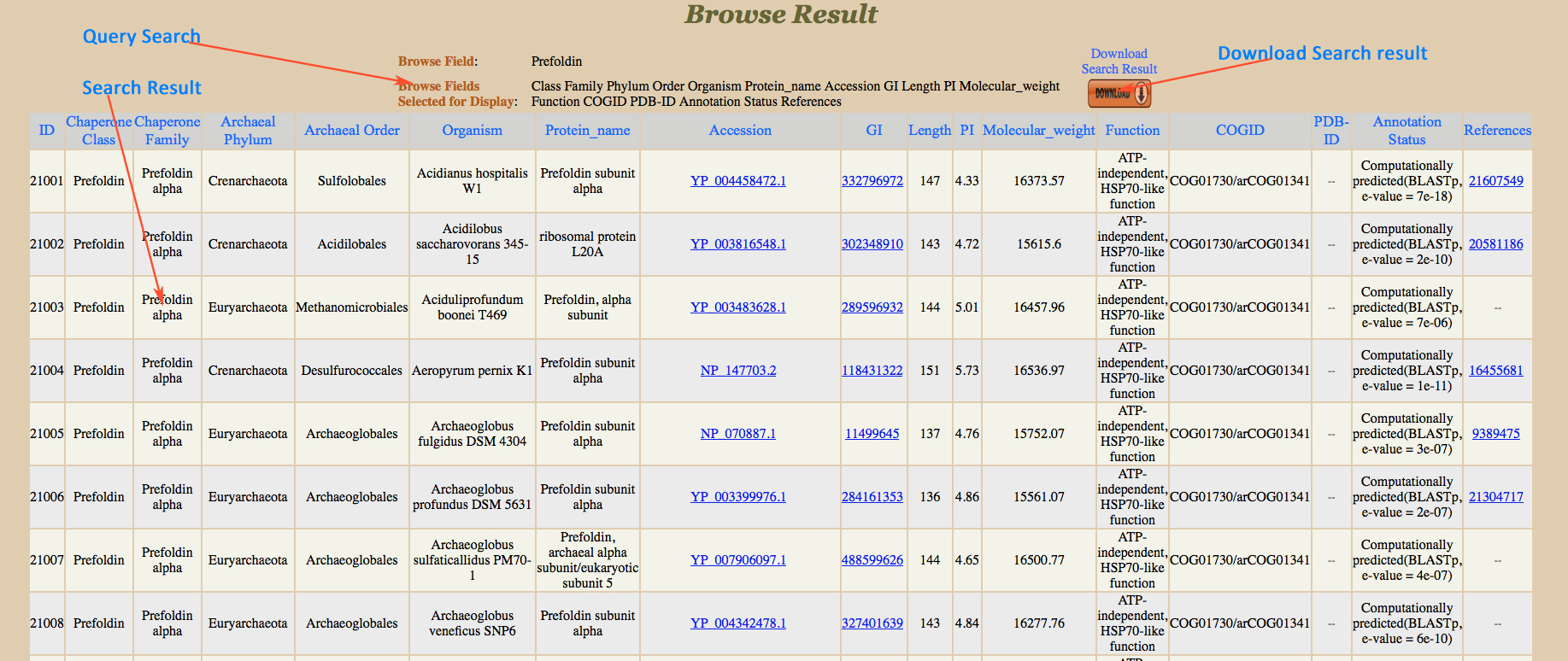 |
BLAST Search |
Blast-Search page assists the user to run BLAST search against the CrAgDb database. The server provides options to choose different type of databases and expectation value. After submission of job it returns the list of hits similar to the query sequence.
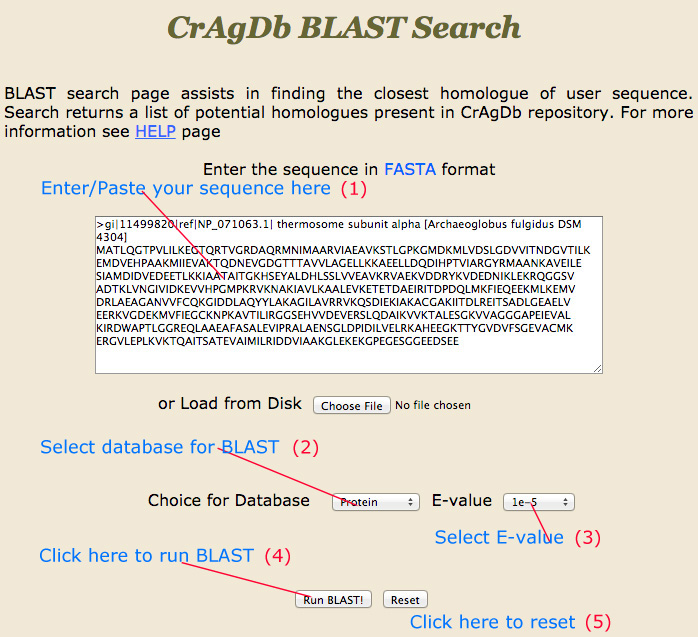 |
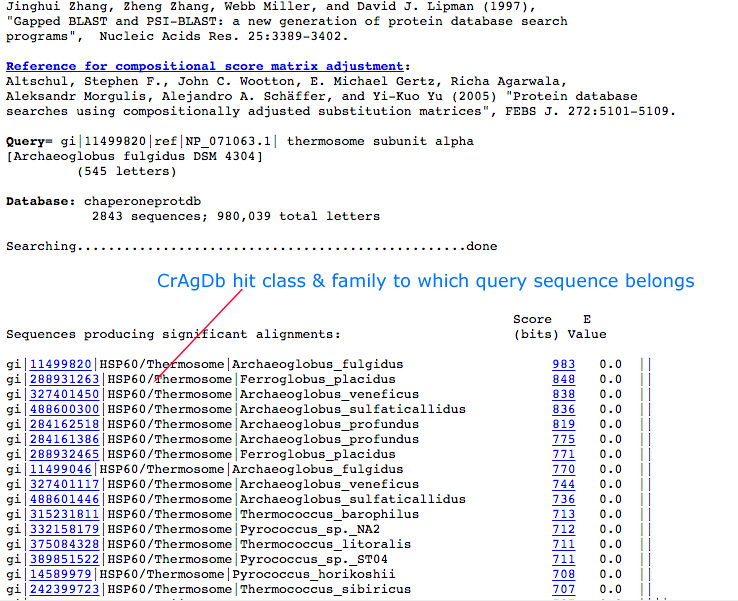 |
Submission and Updation of CrAgDb |
The web page allows user to submit their details on-line by filling a HTML form. However, before including in CrAgDb we will confirm the validity of the submitted information, in order to maintain the quality. Our team is also searching and adding the new chaperone proteins in database from published literature.
Download Information |
This server allows users to download various results used to retrive and analyse during CrAgDb developemnt. All data can be download in .Zip format.
| Select type of information you wish to download | |
|---|---|
| Option | Type of information |
| Protein Files | This allows users to download protein sequence in fasta format. It contains total 4321 sequences. |
| Nucleotide Files | This provides download facilities of Nucleotide sequences in fasta format files. |
| HMM Files | This allows users to download HMMProfile files in .hmm format |
| Alignment Files | This allows users to download Alignment file in .fasta format. |
| Phylogenetic Tree | This allows users to download Phylogenetic Tree in .nwk format. |