CrAgDb
Chaperone Repertoire in Archaeal Genomes

HtpX
(Heat Shock Protein)
HtpX is a heat shock, membrane bound zinc metalloprotease which participates in proteolytic quality control of membrane proteins against misfolded and denatured polypeptides. Under stressful conditions misfolded and misassembeled membrane proteins are produced and these zinc dependent metalloproteases are known to play an essential role in the degradation of unstable membrane proteins (Ref 1; Ref 2).HtpX was first discovered in E.coli as a heat inducible protein with the features of a membrane protein and a metalloprotease (Ref 3) HtpX has the unusual ability to cleave itself autocatalytically in the presence of Zn2+, a divalent cation which helps in the activation of water molecules. The integral membrane protein like HtpX belongs to M48 family of peptidases and contains the conserved motif HEXXH, which is a part of the metal binding site (Ref 4). The sequence and structural analysis showed that HtpX homolog mainly consists of two transmembrane segments, a small N-terminal domain and a large C-terminal domain which includes two hydrophobic regions for the interaction with the membrane (Ref 5). However, some topological studies indicated that the active site of HtpX is located on the cytoplasmic side of the cytoplasmic membrane.
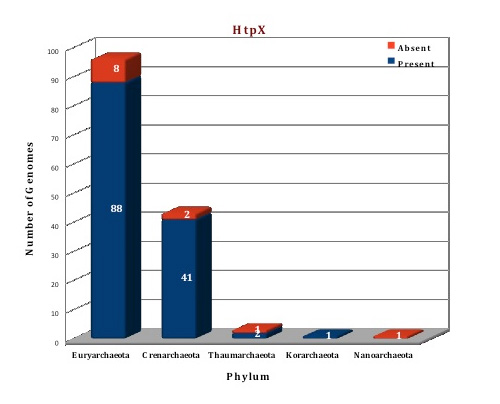
The data of our database shows that the HtpX protein is present in 132 archaeal genomes and absent from 12 genomes (A.boonei, F.acidarmanus, H.paucihalophilus, Halobacterium sp. NRC-1, M.conradii, M.bourgensis, M.kandleri, M.stadtmanae, N.equitans, N.maritimus, P. fumarii, T.cellulolyticus), although two or three copies are present in 48 genomes. All the genomes from Sulfolobus family have three copies of HtpX gene. Two copies are present in some genomes of Halobacteriales, Methanosarcinales and Thermoproteales. The crystal structure of HtpX from Saccharomyces mikatae (4IL3) (Ref 6) and Vibrio parahaemolyticus (3CQB) is available but HtpX from Picrophilus torridus has very little sequence similarity with these proteins, so Phyre2 server (Ref 7) was used for generating the structure on the basis of the template from Saccharomyces mikatae.
|
|
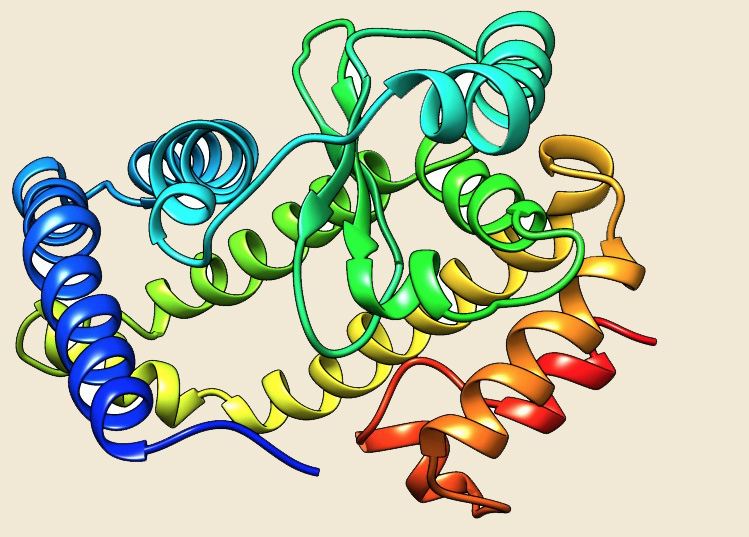
Figure: The modeled structure of HtpX from Picrophilus torridus. |
|
|
|
Figure: Distribution of HtpX chaperone proteins in 144 archaeal genomes. |
 |
References:
1. Proteolytic activity of HtpX, a membrane-bound and stress-controlled protease from Escherichia coli.![]()
2. Cellular functions, mechanism of action, and regulation of FtsH protease.![]()
3. Isolation, characterization, and sequence of an Escherichia coli heat shock gene, htpX.![]()
4. Evolutionary families of metallopeptidases.![]()
5. The Cpx stress response system of Escherichia coli senses plasma membrane proteins and controls HtpX, a membrane protease with a cytosolic active site.![]()
6. Structure of the integral membrane protein CAAX protease Ste24p.![]()
7. Protein structure prediction on the Web: a case study using the Phyre server.![]()
