CrAgDb
Chaperone Repertoire in Archaeal Genomes

Chaperonin-Group I
(Cpn60 & Cpn10)
Chaperonins (commonly also known as Hsp60) are found in all three domains of life. On the basis of their evolutionary lineage, these proteins can be divided into two groups: Group I and Group II. In general, Group I chaperonins are found in eubacteria and in mitochondria and chloroplast of eukaryotes. Group II chaperonins are ubiquitously found in eukaryotic cytosol and also in archaea. However, few archaeal genomes have been reported to have Group I chaperonin homologs also, thought to have been acquired through horizontal gene transfer (Ref 1, Ref 2). Group I chaperonins are also known as Cpn60 which use members of Cpn10/Hsp10 family as co-chaperones. In Bacteria, Cpn60/Hsp60 is known as GroEL, whereas its 10kD co-chaperone counterpart Cpn10 is known as GroES.
The structure, mechanism of action and cellular role of Group I chaperonins is much more widely investigated as compared to the members of Group II. Chaperonins of both group I and II have a characteristic double ring assembly, where each subunit contains three domains: an "equatorial" domain that contains an ATP-binding site, an "apical" domain for substrate binding having a large hydrophobic surface patch in the form of helix-turn-helix motif and an "intermediate" domain that connects the equatorial and apical domains through flexible hinges (Ref 3, Ref 4). The Group I chaperonins (Cpn60) exist as ~800kDa homotetradecamers, which are arranged in two stacked heptameric rings. The cofactor Cpn10 forms a single homoheptameric ring that associates with end of the Cpn60 cylinder, forming an enclosure for the unfolded protein substrate.
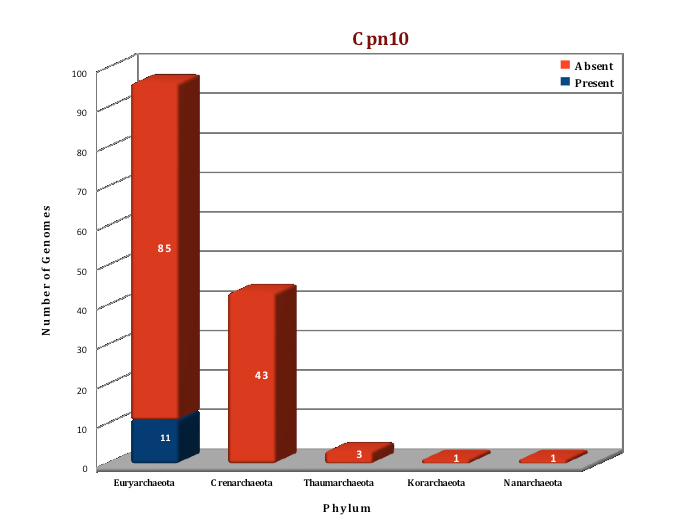
Preliminary data analysis of our database suggest that only 11 archaeal genomes out of the currently sequenced 144 genomes have a homolog of the Group I chaperonins. Cpn60 and its cofactor Cpn10 are present in seven species of Methanosarcinales, three species of Methanomicrobiales and one species of methanococcales. No structure of any archaeal Group I chaperonin is yet present in PDB database, so we have modeled the structure of Cpn 60 and Cpn10 of Methanosarcina acetivorans using the Thermus thermophilus homologs as templates (4V4O (Ref 5) and 1WNR (Ref 6)), respectively.
Cpn60 |
|
|
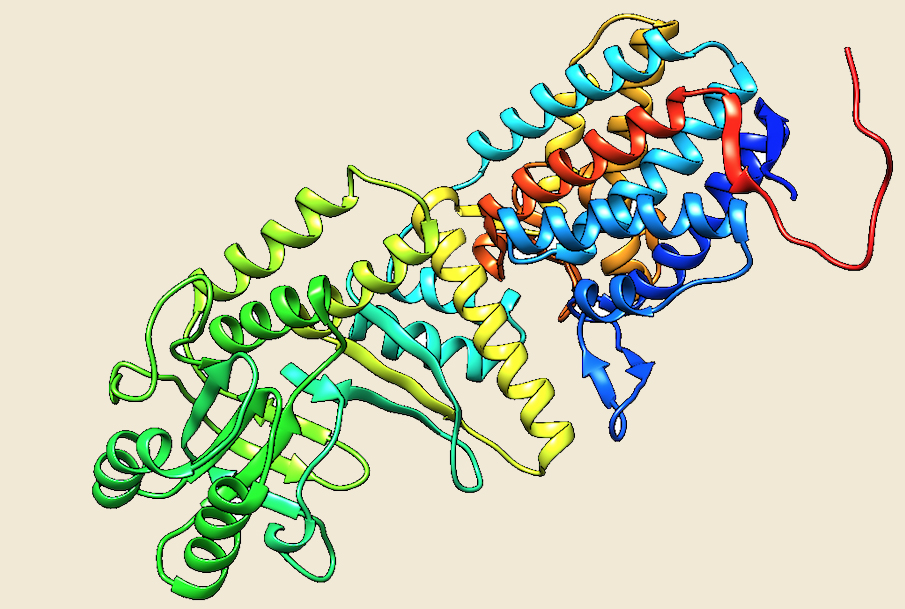
Figure: The modeled structure of Cpn60 from Methanosarcina acetivorans. |
|
|
|
Cpn10 |
|
|
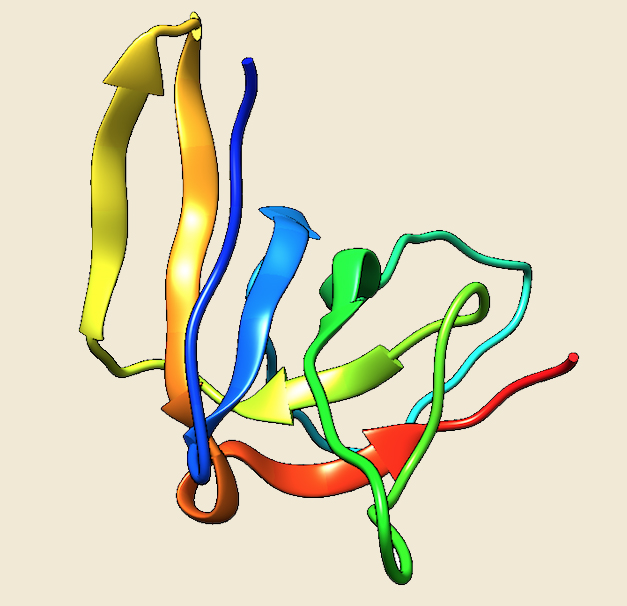
Figure: The modeled structure of Cpn10 from Methanosarcina acetivorans. |
|
|
|
Figure: Distribution of chaperonin-Group I proteins in 144 archaeal genomes. |
 |
 |
References:
1. Coexistence of group I and group II chaperonins in the archaeon Methanosarcina mazei![]()
2. The chaperonins: perspectives from the Archaea.![]()
3. Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT.![]()
4. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex.![]()
5. Crystal structure of the native chaperonin complex from Thermus thermophilus revealed unexpected asymmetry at the cis-cavity![]()
6. Numoto N, Kita A, Miki K. Crystal structure of the Co-chaperonin Cpn10 from Thermus thermophilus HB8. Proteins. ![]()
