β-LacFamPred: HMM based prediction and annotation tool for β-lactamase families
How to Use (Tutorial)
β-LacFamPred predict the beta-lactamase families into proteins/gene sequences.
Tips to use β-LacFamPred Tool
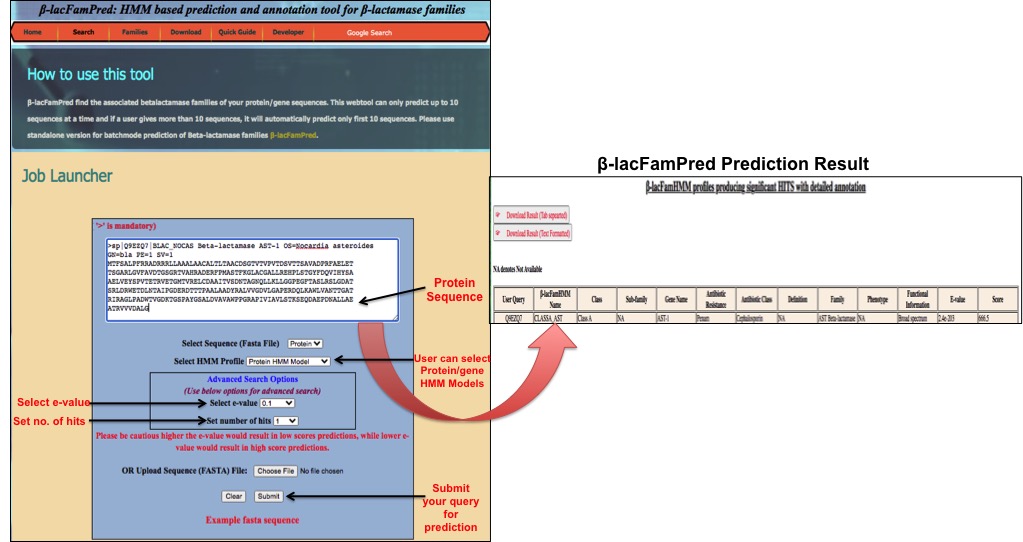
User(s) have option to cut and paste sequences in the text box or to directly upload the sequence file in the search page. Users need to select from the drop box (protein/Gene) sequence accordingly. All sequences must be in the FASTA format. β-LacFamPred scans protein sequences as well as gene sequences provided in fasta format. To ensure that every sequence has a unique ID and it should be in correct format. Special characters (including white space, dot and comma) will be removed from the sequences. However, if a sequence contains illegal letters (e.g., numbers) and non-standandred nucleotide, the sequence will not be processed. β-LacFamPred also provide to users an option to select HMM profile of AR gene (protein ARG HMM i.e. pARGhmm or Nucleotide ARG HMM i.e. nARGhmm). If user submits gene sequence and select pARGhmm then the query will be first translated to protein sequences and then scanning will be proceeded. Users have an option to scan directly without translation uisng nARGhmm. For protein sequences users has to select directly protein and pARGhmm from drop box (protein) & (pARGhmm). β-lacFamPred can process upto 100 sequences at a time. If user submits more than 100 sequences, β-LacFamPred will predict only first 100 sequences in one go. Input
For each submitted job (gene/protein sequence), the tool will produce scanning result in tabular form. First column of result table shows the user query ID, contains similarity score and e-value of query against user query and tool will also provides detailed annotation of query (gene name, description, class name, family name, antibiotic class, resistance mechanism, phenotype, functional information etc). The result can be downloaded in text format as well as in the tab separated format. Here is a sample of scanning done in response to a query protein showscomplete annotation with e-value score and last column represent the prediction score against query sequence. On the basis of e-value and score user can optimize/select the hit.