

| BlaPred: is a three tier prediction system specially trained for classifying and predicting β-lactamase into their respective class and sub-class according to the Ambler's classification. The prediction server is based on classic or Type-1 pseudo amino acid composition based svm model and have been trained and tested using leave one out cross-validation technique on the dataset of 3298 proteins (298 β-lactamase & 3000 non-β-lactamase proteins). |
Prediction Schema |
| The basic architecture of the proposed scheme for predicting and classifying β-lactamase has been shown in the figure given below. The proposed strategy comes in three stages. At the first stage, its check weather the protein is β-lactamase or not. If it is a predicted as a β-lactamase, then at the second step it predicts to which of the four β-lactamase class it belongs. If it is predicted as class B β-lactamase (Metallo β-lactamase) then the sub-classes of class B will be predicted at third-tier of prediction. |
Complete workflow of whole prediction algorithm |
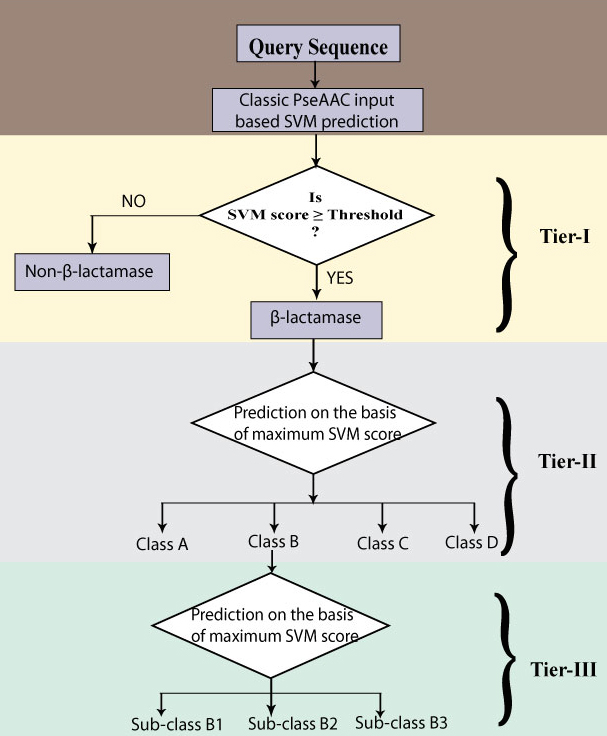 |
Support vector machine |
The support vector machine is the kernel-based classifier based on statistical and optimizing theory. The SVM always seeks global hyperplane to maximize the separating margins between the both classes of examples in training set and avoid overfitting.The SVM is attractive to biological analysis due to its ability to handle noise and large dataset. SVM have shown to performed better in the prediction of various states of proteins such as subcellular classification, enzyme classification etc. and has achieved remarkable success. In the present study, we use SVMlight to predict and classify β-lactamase (http://www.cs.cornell.edu/People/tj/svm_light/) |
Performance measures |
Leave One Out cross-validation |
| The evaluation of accuracy of prediction method is necessary to estimate the performance of a method. The Leave One Out Cross-Validation (LOOCV) approach of learning was employed to estimate the performance accuracy of the prediction methods. The performence of the method is evaluated by using the Senstivity , Specificity, specificity and Accuracy.The results were obtained by averaging the results over the testing subsets. i.e. equal to the number of examples in the training data. The formula's of sensitivity specificity, MCC calculations are shown below: |
| Sensitivity = (TP / (TP+FN))*100† |
| Specificity = (TN / (TN+FP))*100 |
| Accuracy = (TP+TN / (TP+FP+TN+FN))*100 |
| MCC = (TP * TN) - (FP*FN) / ÷(TP+FN)* (TP+FP)*(TN+FP)*(TN+FN). |
| Where†TP†and†TN†are correctly predicted as β-lactamase and non-β-lactamase respectively.†FP†and†FN†are wrongly predicted β-lactamase and non-β-lactamase respectively. |
Copyright © 2016 -Department of Biophysics,University of Delhi South Campus,New Delhi India
Contact to Dr.Manish Kumar for Bugs/comments at Email:manish at south dot du dot ac dot in