

We provide both positive and negative dataset that we used to developed different SVM models.We are providing β-lactamase and non-βlactamase so that any user can download it for research purpose.
Click HERE to get the positive dataset used in training SVM module in BlaPred.
Click HERE to get the negative dataset used in training SVM module in BlaPred.
Click HERE to get the independent dataset used in BlaPred.
Click HERE to get the Supplementery tables of BlaPred.
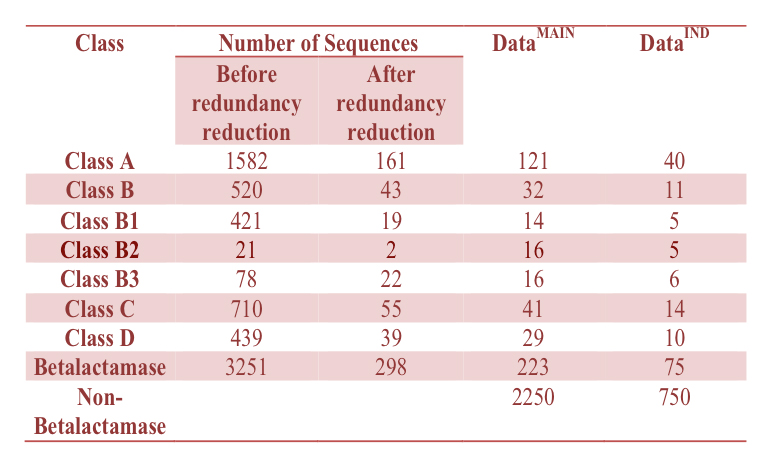
The training/testing dataset contains 298 proteins classified into four classes (class A B C D) and three subclass (B1,B2 and B3) according to the information available. None of the proteins has >60% sequence identity. The Uniprot Knowledgebase entry ID's of these 298 protein sequences are available for download. (Click here)
 |
Additional information on various modules developed...!! |
Amino Acid Composition: - |
The information of a protein can be encapsulated in a vector of 20 dimensions using amino acid composition of the protein. The composition was used as input in this study, which provides the global information of protein features in the form of fixed length vector. The amino acid composition is the fraction of each amino acid type within a protein. The fractions of all 20 natural amino acids were calculated by using the following equation: | 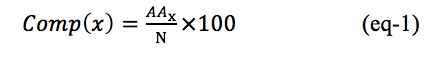 |
| Here Comp (x) is amino acid composition of residue type AAx and N is the total number of amino acids in protein sequences. |
Pseudo Amino Acid Composition: - |
| Type-1 or Classic Pseudo amino acid composition was used to utilize the sequence order information in prediction and classification of β-lactamase.The SVM module predicted an overall accuracy of 93.57% which was higher than other composition based SVM module i.e Type-2 or Amphiphilic PseAAC and amino acid composition. The results obtained after leave one out cross-validation for classic pseudo amino acid composition are shown below: |
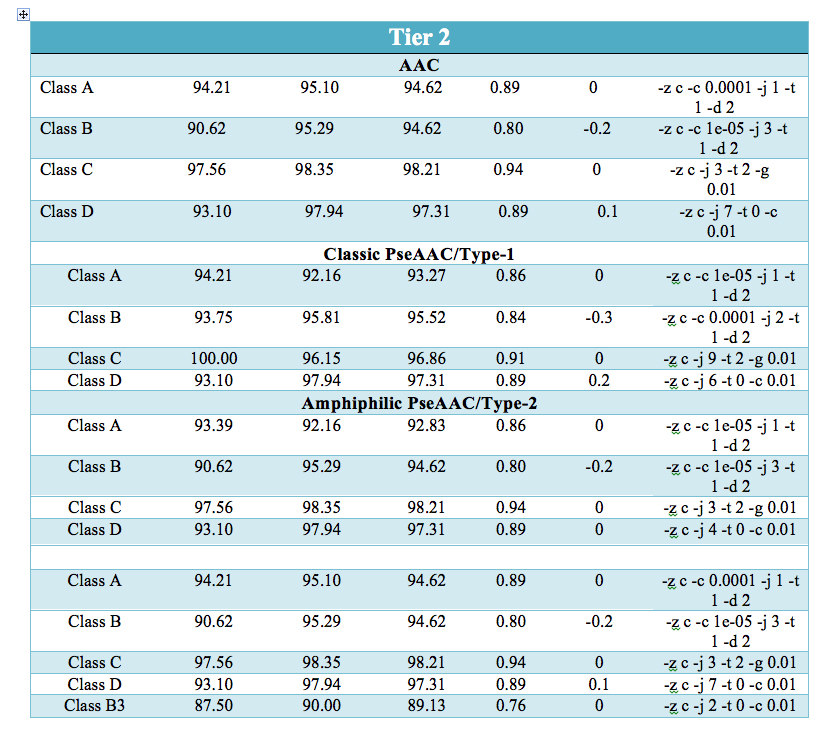
Prediction performace of SVM modules at all the three levels: - |
| The prediction results are shown in user-friendly format. The prediction results provides information at three separate level for classifying β-lactamase i.e. the predicted sequence belong to β-lactamase class or not (Tier-1). At second(Tier-2) and third (Tier-3) level, it predicts sequence belongs to which of the four class(A,B,C,D) and three subclass(B1,B2,B3) respectively. | 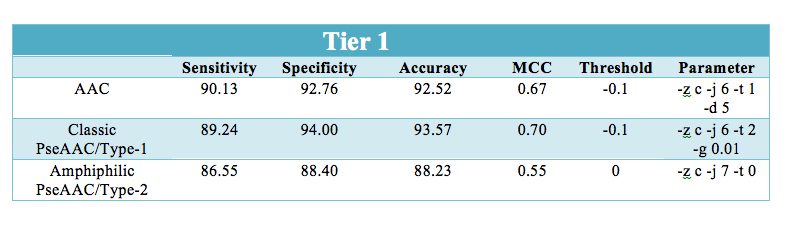 |
 |
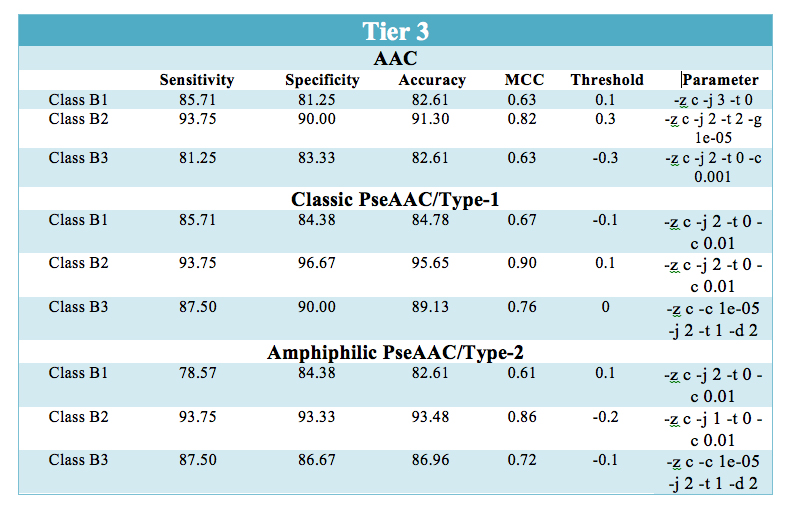 |
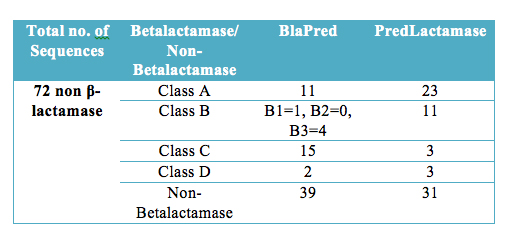
Table 3. Webserver wise comparative performance on benchmark independent dataset. |
| Prediction performance on benchmarking dataset were also checked to confirm whether BlaPred can identify homologous proteins of β-lactamase as non-β-lactamase. BlaPred correctly predicted 39 DD-peptidases as non-β-lactamase out of total 72. PredLactamase could able to correctly predict only 31 DD-peptidases as non-β-lactamase. |
 |
Copyright © 2016 -Department of Biophysics,University of Delhi South Campus,New Delhi India
Contact to Dr.Manish Kumar for Bugs/comments at Email:manish at south dot du dot ac dot in