CrAgDb
Chaperone Repertoire in Archaeal Genomes

PDIases
(Protein Disulfide Isomerases)
Protein disulfide isomerase (PDI) is a well know enzyme that can catalyse the rearrangement of disulphide bonds in proteins, and has been reported to have chaperone-like activity at high concentrations (such as inhibition of aggregation). The formation of disulfide bridge is a common structural element in proteins that provides covalent links between cysteine residues, and is generally known as a rate limiting step in protein folding (Ref 1; Ref 2). A significant fraction of proteins in the cytoplasm of thermophilic and hyperthermophilic archaea exhibit disulfide bonds and it is widely considered that these bonds may contribute to the thermostability of these proteins (Ref 3). However very little structural and functional information is available on proteins that are capable of catalyzing this reaction in archaea, such as PDIases.
Protein disulfide isomerase has two catalytic sites, one situated near the NH2 terminus and the other near the COOH terminus. Protein disulphide isomerases are traditionally categorized into different families like thioredoxin, glutaredoxin and many other disulphide bond forming proteins which are characterized by an active site having CXXC sequence motif (Ref 4; Ref 5). Interestingly, all the proteins of this family have two potential active sites with the conserved motif CXXC and both the sites appear to be necessary for disulphide isomerase activity.
The structural and functional studies have revealed that the archaeal forms of this protein show oxidative and reductive activity of disulphide bonds, which is mediated by thiol disulphide exchange reaction between the active site cysteines of the enzyme and cysteine in the target protein (Ref 6). It is assumed that an interplay of thermodynamic and kinetic effects including structure of the active site may play an important role in the regulation of disulphide bond formation, in analogy to the situation seen in protein disulphide oxidoreductases. The thioredoxins and glutaredoxins have been well studied in prokaryotes and eukaryotes, but to date very little structural and functional information is available on PDIs from archaea.PDIases with multiple thioredoxin/glutaredoxin domains within a single polypeptide are known in eukaryotes but in archaeal organisms single domain proteins are present.
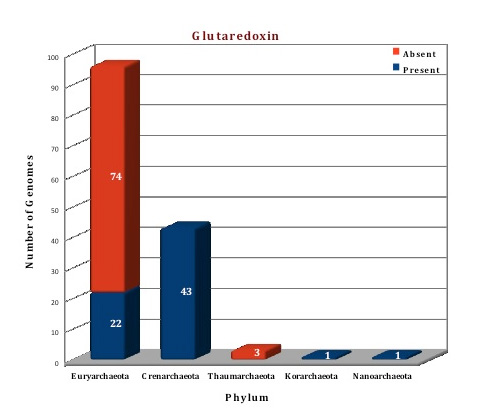
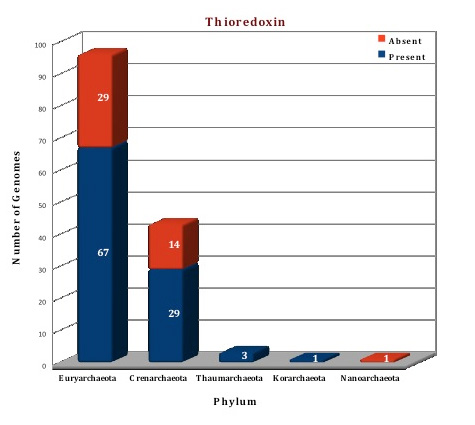
The data of our database, suggests that both the thioredoxin and glutaredoxin proteins are present in archaeal genomes. Thioredoxin is found in 100 genomes (often in more than two copies) and is totally absent from 44 genomes. Glutaredoxin was observed in 67 genomes but is absent from 77 genomes, although two copies of this protein are present in 15 genomes(M.cuprina, M.sedula,S.acidocaldarius, S.islandicus HVE10/4,S.islandicus L.D.8.5, S.islandicus L.S.2.15, S.islandicus M.14.25, S.islandicus M.16.27, S.islandicus M.16.4, S.islandicus REY15A, S.islandicus Y.G.57.14, S.islandicus Y.N.15.51, S.solfataricus 98/2, S.solfataricus P2, S.tokodaii). The crystal structures of both the proteins are available from archaeal kingdom. The structure of thioredoxin from Sulfolobus solfataricus (3HHV) (Ref 7) and thioredoxin like protein from Sulfolobus tokodaii (2E0Q) (Ref 8) is available. We have modelled the structure of thioredoxin from Picrophilus torridus by using 3HHV from Sulfolobus solfataricus as template. The crystal structure of Glutaredoxin and glutaredoxin like proteins are available from Pyrococcus horikoshii (1J08), Pyrococcus furiosus (1A8L) (Ref 9), Archaeoglobus fulgidus (3IC4), Methanosarcina mazei (3NZR). Based on the template 1JO8, the structure of glutaredoxin from Picrophilus torridus was modelled.
Glutaredoxin |
|
|
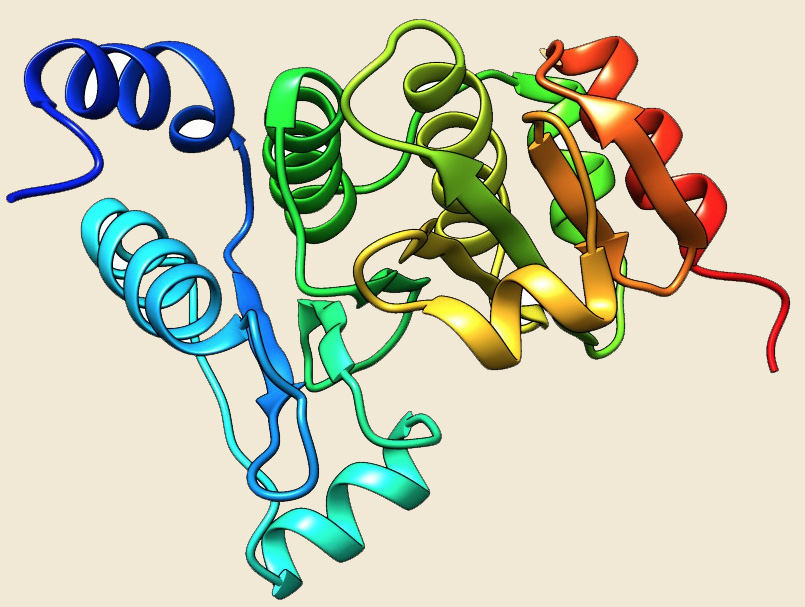
Figure: The modeled structure of PDIases from Picrophilus torridus. |
|
|
|
Thioredoxin |
|
|
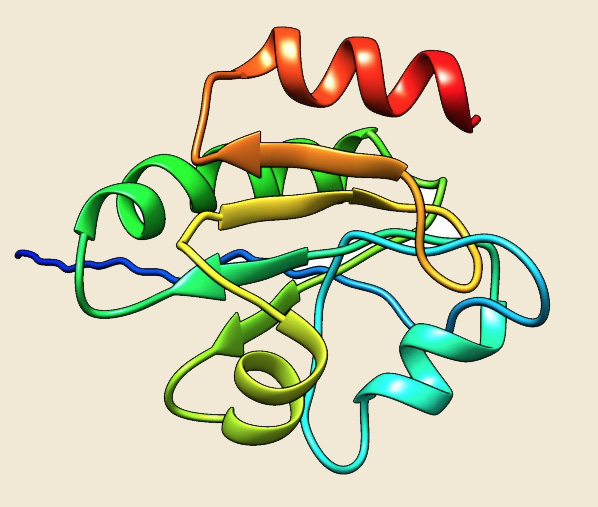
Figure: The modeled structure of PDIases from Picrophilus torridus. |
|
|
|
Figure: Distribution of PDIases chaperone proteins in 144 archaeal genomes. |
 |
 |
References:
1. Formation of disulfide bonds in proteins and peptides.![]()
2. Limited tendency of alpha-helical residues to form disulfide bridges: a structural explanation.![]()
3. Protein disulfides and protein disulfide oxidoreductases in hyperthermophiles.![]()
4. Protein disulfide bond formation in prokaryotes.![]()
5. Thioredoxins and glutaredoxins as facilitators of protein folding.![]()
6. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein.![]()
7. The dimeric structure of Sulfolobus solfataricus thioredoxin A2 and the basis of its thermostability.![]()
8.Crystal structure of thioredoxin domain of ST2123 from thermophilic archaea Sulfolobus tokodaii strain7.![]()
9. A protein disulfide oxidoreductase from the archaeon Pyrococcus furiosus contains two thioredoxin fold units.![]()
