CrAgDb
Chaperone Repertoire in Archaeal Genomes

PPIases
(Peptidyl Prolyl cis-trans Isomerase)
Peptidyl prolyl cis-trans isomerases (PPIases) is a class of enzymes that promote the rotation of peptidyl prolyl bond and shows chaperone like protein folding activity. PPIases traditionally comprise of three non-homologous proteins: FKBPs (FK506 binding proteins), Cyclophilins (cyclosporin A binding protein) and Parvulins, all of which share the ability to catalyze the bond between the cis and trans forms of a proline residue (Ref 1). FKBP's and Cyclophilins have earlier been analyzed in several archaeal genomes and assigned a function related to chaperone activity. However, archaeal parvulin sequences have became available only in last few years as more and more new genomes have been sequenced, thus their cellular function has not yet been studied in detail yet. Archaeal parvulins comprise of a single domain with a molecular weight of about 10 kDa but single domain parvulins are absent from eukaryotic genomes. Archaeal FKBP have been classified into two groups, Long type (26-33 kDa) FKBP and Short type FKBP (16-18 kDa). Long type FKBP consist of two domains, N-terminal domain and C-terminal domain but short type FKBP contain only one domain which is homologous to N-terminal domain of long type FKBP (Ref 2). It has been reported that both the types of FKBP have chaperone like activity (Ref 3, Ref 4, Ref 5). The cyclophilin type PPIases act as target for the immunosuppressant cyclosporine. While PPIase activities of cyclophilins and FKBP's are generally inhibited by their corresponding immunosuppressants, cross inhibition is not observed, thus suggesting that they differ in their mode and mechanism of action.
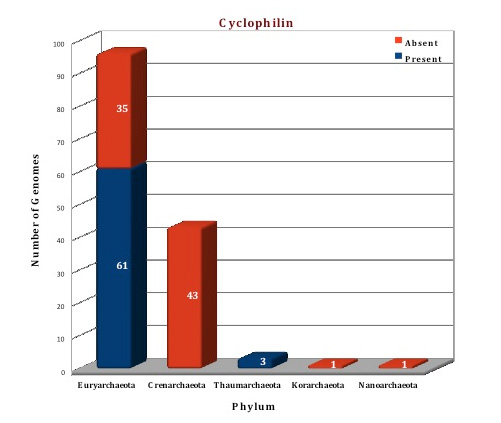
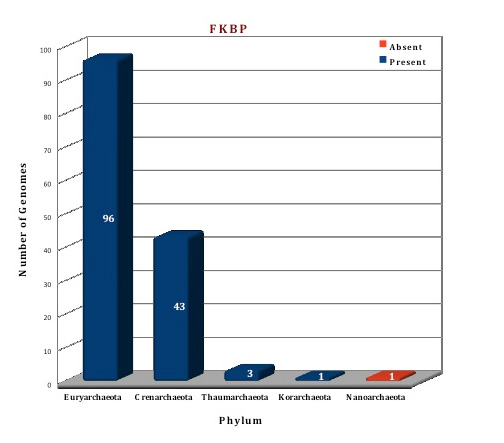
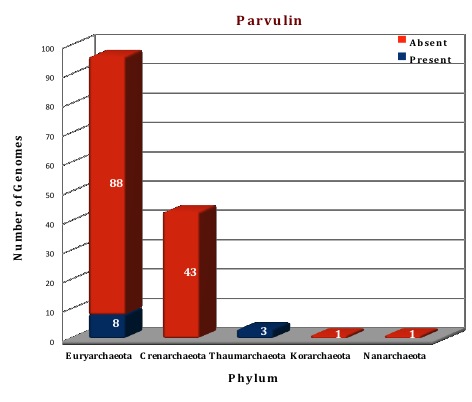
Preliminary analysis of the data in our database suggests that FKBP is the only ubiquitous protein of PPIases family present in all the archaeal (144) genomes except in Nanoarchaeum equitans (which can be considered as a special case because of its dependency on Ignicoccus). All the archaeal genomes do not have both types of FKBP, only 83 genomes are having both the long and the short type, but the other 60 genomes have only long type FKBPs. Cyclophilins and parvulins are absent from all the species of phylum Crenarchaeota, Korarchaeota and Nanoarchaeota, the genomes for which have been sequenced till date. Even in Phylum Euryarchaeata, several orders like Archaeoglobales, Methanopyrales and Methanococcales do not have the cyclophilin and parvulin present. Parvulin protein can only be found in the phylum Thaumarchaeota, and a few species of Euryarchaeota (Methanomicrobiales and Methanosarcinales), thus present in a total of eleven genomes (one protein per genome).
Two crystal structures of FKBP are currently available from the archaeal species M.jannaschii (3PRB) (Ref 6) and M.thermolithotrophicus (1IX5) (Ref 7) and one structure of archaeal parvulin is available from Cenarchaeum symbiosum (2RQS) (Ref 8) but no structural information is available on cyclophilins from archaea. The structures of long type FKBP and cyclophilin proteins from Picrophilus torridus were modelled based on the template M.jannaschii (3PRB) and Homo sapiens (2X7K) (Ref 9) for FKBP and cyclophilin respectively.
Cyclophilin |
|
|
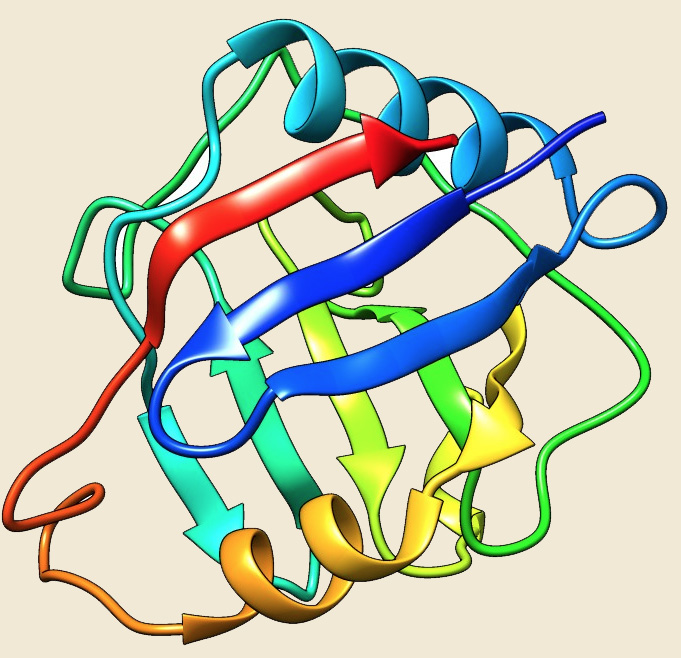
Figure: The modeled structure of Cyclophilin from Picrophilus torridus. |
|
|
|
FKBP |
|
|
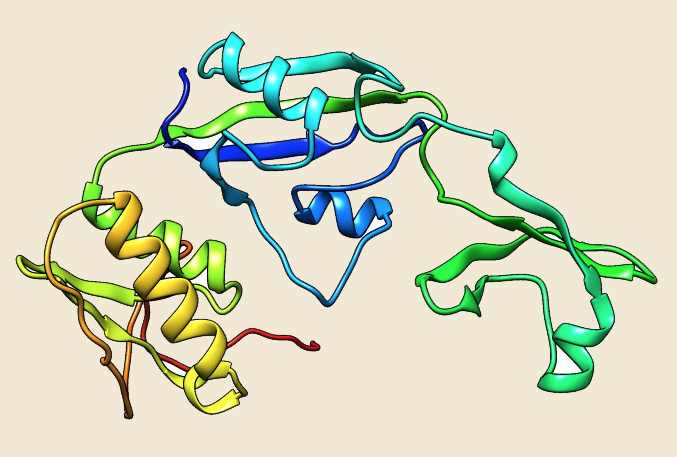
Figure: The modeled structure of FKBP from Picrophilus torridus. |
|
|
|
Parvulin |
|
|
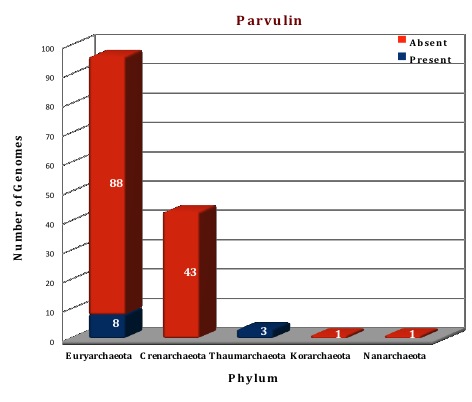
Figure: The modeled structure of Parvulin from Nitrosopumilus maritimus. |
|
|
|
Figure: Distribution of PPIases chaperone proteins in 144 archaeal genomes. |
 |
 |
 |
References:
1. Archaeal peptidyl prolyl cis-trans isomerases (PPIases) update 2004.![]()
2. Archaeal peptidyl prolyl cis-trans isomerases (PPIases).![]()
3. The 28.3 kDa FK506 binding protein from a thermophilic archaeum, Methanobacterium thermoautotrophicum, protects the denaturation of proteins in vitro.![]()
4. FK506-binding protein of the hyperthermophilic archaeum, Thermococcus sp. KS-1, a cold-shock-inducible peptidyl-prolyl cis-trans isomerase with activities to trap and refold denatured proteins.![]()
5. FK506 binding protein from the hyperthermophilic archaeon Pyrococcus horikoshii suppresses the aggregation of proteins in Escherichia coli.![]()
6. Structural analysis of protein folding by the long-chain archaeal chaperone FKBP26.![]()
7. Three-dimensional solution structure of an archaeal FKBP with a dual function of peptidyl prolyl cis-trans isomerase and chaperone-like activities.![]()
8. Structure and dynamics of the first archaeal parvulin reveal a new functionally important loop in parvulin-type prolyl isomerases.![]()
9.The crystal structure of PPIL1 bound to cyclosporine A suggests a binding mode for a linear epitope of the SKIP protein.![]()
