CrAgDb
Chaperone Repertoire in Archaeal Genomes

AAA+ ATPases
(ATPases Associated with a variety of cellular Activities)
The AAA family (ATPases Associated with a variety of cellular activities) are a group of oligomeric, energy dependent proteins whose major activity seems to be the unfolding and degradation of various cellular proteins and protein complexes (Ref 1; Ref 2). AAA+ superfamily proteins are ubiquitous enzymes found in all biological kingdoms, with P-loop NTPases forming the largest protein superfamily which acts as molecular chaperones. Structurally, AAA+ ATPase proteins are form hexameric rings with an N-terminal domain (N), thought to mediate substrate recognition, followed by one or two extended ATPase domains (D1 or D2), both containing AAA modules. The high degree of structural conservation is observed in central ATPase domain of about 200-250 amino acid region (responsible for ATP binding and hydrolysis) among all the AAA+ superfamily members but the N-terminal domains are divergent (Ref 3; Ref 4 ; Ref 5). The N-domain appears to be flexibly linked to the core complex and the structural analysis of this domain reveals its composition in two subdomains: an N-terminal α/β core subdomain and a C-terminal α-helical subdomain (Ref 4). Other than that, the proteins of this superfamily show three consereved motifs, Walker A (GxxGxGKT), Walker B (hhhhDE) and Sequence region for homology (SRH) which are responsible for ATP binding and hydrolysis.
The archaeal organisms contain several proteins from AAA+family that are likely to have chaperone like activities in the folding and unfolding of proteins and protein complexes, e.g Lon proteases and the Cdc48 homologue VAT. Lon is the first known ATP dependent protease, also known as protease La, which is highly conserved in all the three domains of life (Ref 6; Ref 2; Ref 7). Mechanistically, Lon proteases is similar to other ATP-dependent proteases in ATP binding and hydrolysis, because of a significant degree of sequence homology (Ref 8). Vat (VCP-like ATPases) is the member of CDC48/p97 subfamily. CDC48 of Saccharomyces cerevisiae (Ref 9) and p97 of vertebrates, also known as the valosine-containing protein (VCP), (Ref 10) are well-characterized members of this subfamily. Vat forms hexameric complexes indicating that the N-domain is not required for the dimerization and is rather flexible in position. D1 and D2 domains have the same folds and are believed to sit on top of each other (Ref 11).
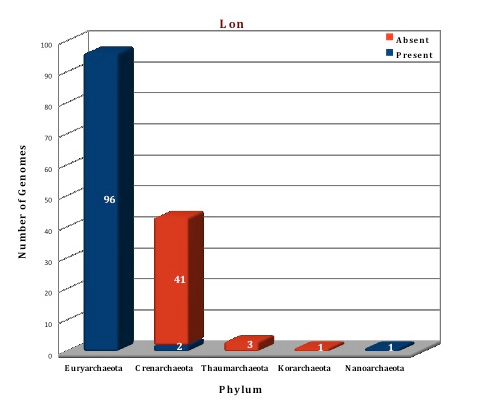
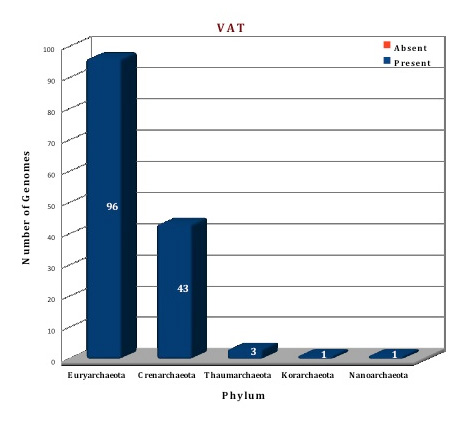
As per the data of our database, AAA+ ATPase superfamily constitutes a huge class of chaperone proteins in archaeal organisms. VAT protein is a ubiquitous family of this superclass and present in all the archaeal genomes. More than three copy numbers are present in almost all the genomes. Whereas Lon is present in 99 genomes and is totally absent from the phylum Crenarchaeota, Thaumarchaeota and Korarchaeota. The second Lon homolog, annotated as Lon-2, is found in 14 genomes (A.boonei F.acidarmanus, Methanobacterium sp. AL-21, Methanobacterium sp. Swan-1, M.marburgensis, M.thermautotrophicus, M.fervidus, N.Pellirubrum, P.torridus, T.acidophilum, T.volcanium, T.archaeon BRNA-1) and consists of AAA+ domain only and lacks the C-terminal protease domain present in Lon. The cellular function of this protein is still not clear. Three Lon structures (1XHK, 1Z0E, 3K1J) (Ref 12; Ref 13, Ref 14) from archaeal organisms M.jannaschii, A.fulgidus and T.onnurineus respectively are available in the PDB database and one crystal structure of VAT from A.hospitalis (4LCB) (Ref 15) is currently available. We have modelled the structure of Lon protein from Picrophilus torridus byusing the template of T.onnurineus and the Vat protein was modeled using the template 3CF1 from Mus musculus (Ref 16).
LON |
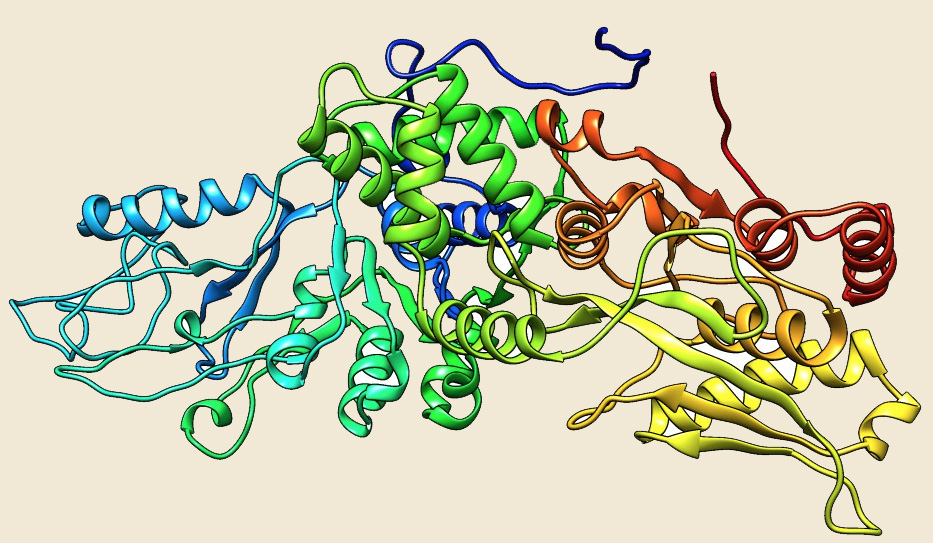
Figure: The modeled structure of AAA+ATPases from Picrophilus torridus. |
||
|
|
VAT |
|
|
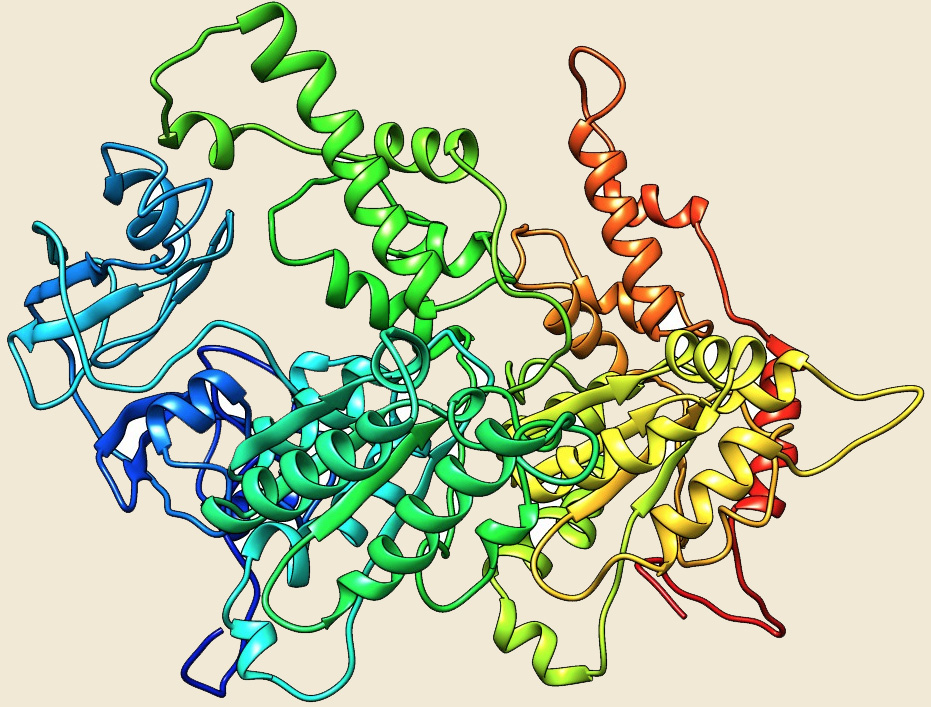
Figure: The modeled structure of AAA+ATPases from Picrophilus torridus. |
|
|
|
Figure: Distribution of AAA+ATPases chaperone proteins in 144 archaeal genomes. |
 |
 |
References:
1. AAA proteins: in search of a common molecular basis. International Meeting on Cellular Functions of AAA Proteins.![]()
3. AAA+ proteins: have engine, will work.![]()
4. AAA+ superfamily ATPases: common structure--diverse function.![]()
5. Phylogenetic analysis of AAA proteins.![]()
6. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes.![]()
7. Evolutionary history and higher order classification of AAA+ ATPases.![]()
8. Functional mechanics of the ATP-dependent Lon protease- lessons from endogenous protein and synthetic peptide substrates.![]()
9. Yeast cell cycle protein CDC48p shows full-length homology to the mammalian protein VCP and is a member of a protein family involved in secretion, peroxisome formation, and gene expression.![]()
10. An abundant and ubiquitous homo-oligomeric ring-shaped ATPase particle related to the putative vesicle fusion proteins Sec18p and NSF. ![]()
11. Structure of VAT, a CDC48/p97 ATPase homologue from the archaeon Thermoplasma acidophilum as studied by electron tomography.![]()
12. The active site of a lon protease from Methanococcus jannaschii distinctly differs from the canonical catalytic Dyad of Lon proteases.![]()
13. Atomic-resolution crystal structure of the proteolytic domain of Archaeoglobus fulgidus lon reveals the conformational variability in the active sites of lon proteases.![]()
14. Crystal structure of Lon protease: molecular architecture of gated entry to a sequestered degradation chamber.![]()
15. The oligomeric state of the active Vps4 AAA ATPase.![]()
16. Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change.![]()
